- Home
- :
- All Communities
- :
- Developers
- :
- ArcGIS API for Python
- :
- ArcGIS API for Python Questions
- :
- Re: ArcGIS GeoAccessor Overlay Erase
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
ArcGIS GeoAccessor Overlay Erase
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
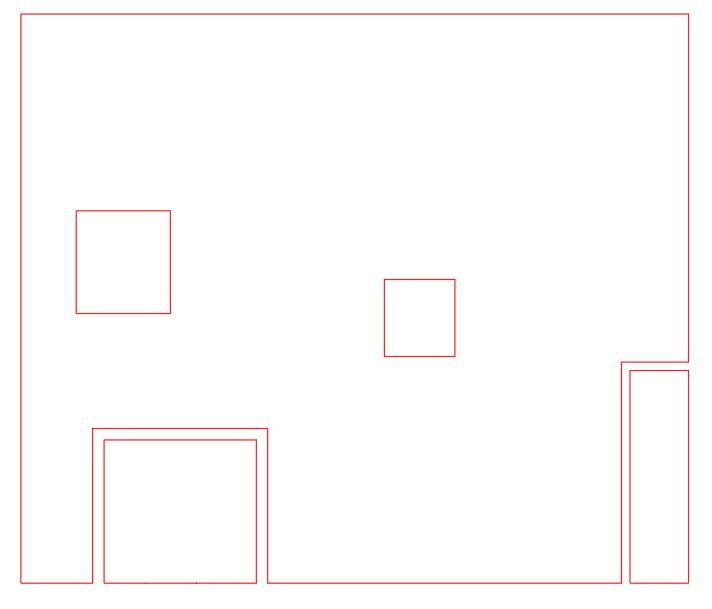
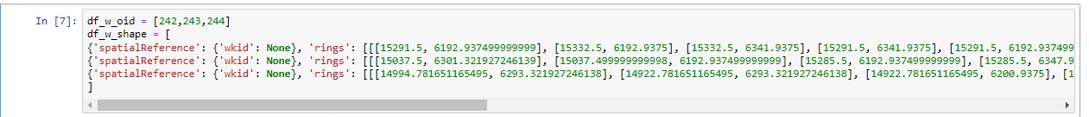
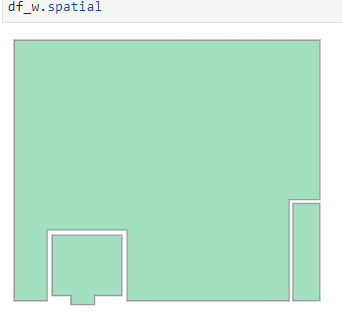
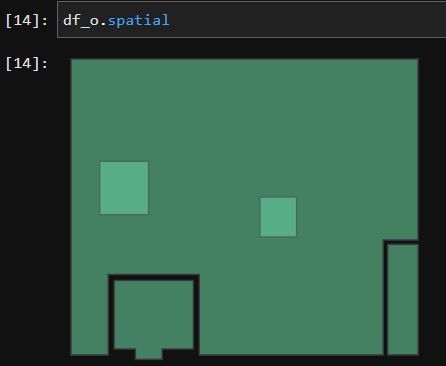
I have two SEDFs. df_w has 3 polygons. df_h has 2 polygons both within one of the df_w polygons. Ultimately I need to cut holes in df_w with the df_h polygon. The subject method is throwing an error. I’m missing something about the functionality of the GeoAccessor overlay although it seems straightforward. Anyway, any insight into the issue would be appreciated. Thank you. Tyler
Polygons Visualized:
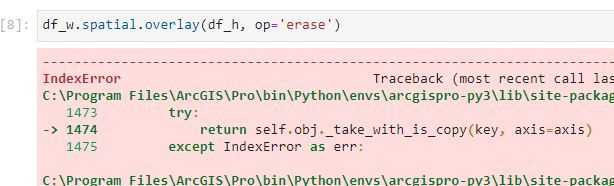
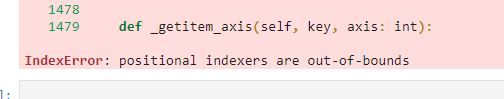
Method and Error:
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
This worked for me
`temp_df.spatial.overlay(temp_df_2,'erase')`
It appears the overlay function is not set up with key word arguments.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
hmm. Removing the 'op=' did not do the trick. Same result. Would you mind posting a screenshot of your dataframes with the index in view. I'm curious to how many features you have and if they share indices. I am sensing overlay is trying to match index to index (from df1 to df2), but not sure.
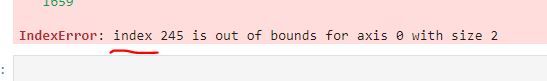
Alternatively, try it with dataframes of different sizes, and different indices. The reason I'm wondering is because when I run op='union' I get this error.
Thank you.
Tyler
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
@DavidAnderson_1701, circling back to see if you have screen shots for your jupiter lab notebook dataframe outputs. Thx. Tyler
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
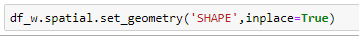
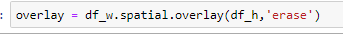
Do the same process for df_h
then
which yields
Short story is it worked for me. Perhaps try recreating the spatial data frames.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
@DavidAnderson_1701 Interesting. Should the two holes be white? What you have shown looks like the holes over the top of the second larger feature. At least you didn't get errors like I got. Let me study this and work on my SEDFs.
Much appreciated.
Tyler
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
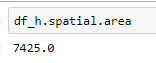
Probably something to do with how I'm displaying the data. I wrestled with how to get the dataframe to show the geometry.
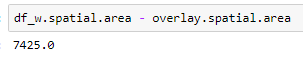
The area values show that the erase did happen.
Not a very satisfying answer I know. Saying it worked for me is an old programmer excuse/cop out. You might double check that the dataframe geometries are indeed polygons using the spatial.area property.
Start a clean Jupyter project, try the code I show to see if that works. Isolate the problem code to just this little bit.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
To display try:
df.spatialExample.
I'll keep wrestling with it, but I'm still getting a positional index error.
Thanks for helping.
Tyler
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Could you post the full traceback stack text. It might be informative to see exactly which module is throwing the error. Is it a Python function? Pandas? Arcgis API?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
@DavidAnderson_1701, I finally made SOME progress. I switched machines and lo and behold, it worked. No error. Original machine has ArcGIS Pro 2.8.3 and the new machine has 2.9.1. I'm wondering if there's a python module bug. Do arcgis/arcpy modules get updated when ArcGIS Pro gets updated? Thanks for all your help troubleshooting. I'll try updating ArcGIS Pro and see what happens.
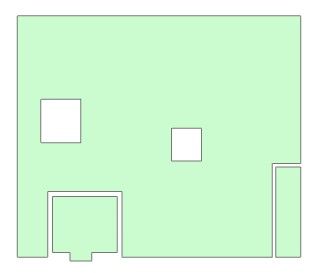
Side note: Jupyter displays holes oddly. They look more like feature over feature, so I checked in ArcGIS Pro and the output feature looks just fine.
Jupyter output feature with holes:
Same SEDF exported to gdb fc and displayed in ArcGIS Pro:
Cheers,
Tyler