- Home
- :
- All Communities
- :
- Developers
- :
- Python
- :
- Python Questions
- :
- Write ArcPy cursor results to a text file in diffe...
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Write ArcPy cursor results to a text file in different folders with a different name based on cursor condition. Define function is not working.Multiprocessing
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Middle section of the code, I separate the attribute value in a different category ("AAA","BBB","CCC") and every category have Ten land-use types, initially, I set Three land use type (LAND_10, LAND_20, LAND_30) for test code. Now I used the update cursor for different land-use type(LAND_10, LAND_20, LAND_30) , and saved the text file to there corresponding Land use folders(Like, Land_10 ....Land_30) under the "AAA" folder. similarly, this function I will use for categories "BBB","CCC" .
Initially, I write this code for category "AAA" but I want to use this function for category "BBB","CCC" by defining a function(uncheck all define function), but is not working. Below is my code please check it.
First of all, I want to reduce my code (Because I have 10 land type initially I write three) and the whole function I want to use as a defined function for a different category.
Another thing is Multiprocessing, Multithreading I want to use this function for the code because of initially my code takes much more time for one processes. In my case how to use the multiprocessing function.
please look at this matter and give me a suggestion to manipulate this code.
import datetime
start = datetime.datetime.now()
print 'start run: %s\n' % (start)
import arcpy ,os ,sys,csv,errno
from arcpy import env
from arcpy.sa import *
import datetime
import re
import glob
import itertools
arcpy.env.overwriteOutput = True
import multiprocessing as mp
print("Number of processors: ", mp.cpu_count())
cellsize = "F:\\DB_test_data\\TEST_RAY\\TEST1\\MOD02HKM_A2017001_0530_NDVI_AA.img"
d1="F:\\DB_test_data\\VAR2\\cldmask\\tt"
CLDMASK = glob.glob(d1 + os.sep + "*.Aerosol_Cldmask_Land_Ocean-Aerosol_Cldmask_Land_Ocean.tif")
CLDMASK.sort()
if CLDMASK is None:
print 'Could not open the CLDMASK raster files'
sys.exit(1)
else:
print 'The CLDMASK raster files was opened successfully'
#print CLDMASK
d2="F:\\DB_test_data\\TEST_RAY\\TEST1"
NDVI = glob.glob(d2 + os.sep + "*A2017001_0530_NDVI_AA.img")
NDVI.sort()
if NDVI is None:
print 'Could not open the NDVI raster files'
sys.exit(1)
else:
print 'The NDVI raster files was opened successfully'
#print NDVI
d3="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND1 = glob.glob(d3 + os.sep + "*A2017001_0530_006_BAND_1.img")
BAND1.sort()
if BAND1 is None:
print 'Could not open the BAND1 raster files'
sys.exit(1)
else:
print 'The BAND1 raster files was opened successfully'
#print BAND1
d4="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND2 = glob.glob(d4 + os.sep + "*A2017001_0530_006_BAND_2.img")
BAND2.sort()
if BAND2 is None:
print 'Could not open the BAND2 raster files'
sys.exit(1)
else:
print 'The BAND2 raster files was opened successfully'
#print BAND2
d4="F:\\DB_test_data\\VAR2\\IDL\\MOD09\\TEST"
BAND3 = glob.glob(d4 + os.sep + "*A2017001_0530_006_BAND_3.img")
BAND3.sort()
if BAND3 is None:
print 'Could not open the BAND3 raster files'
sys.exit(1)
else:
print 'The BAND3 raster files was opened successfully'
#print BAND3
d5="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND4 = glob.glob(d5 + os.sep + "*A2017001_0530_006_BAND_4.img")
BAND4.sort()
if BAND4 is None:
print 'Could not open the BAND4 raster files'
sys.exit(1)
else:
print 'The BAND4 raster files was opened successfully'
#print BAND4
d6 = r"F:\\DB_test_data\\VAR2\\MOD04_l2"
AOD = glob.glob(d6 + os.sep + "*Corrected_Optical_Depth_Land_2-Corrected_Optical_Depth_Land.tif")
AOD.sort()
if AOD is None:
print 'Could not open the AOD raster files'
sys.exit(1)
else:
print 'The AOD raster files was opened successfully'
Scale_factor = float(0.0010000000474974513)
add_offset = float(0.0)
Fill_value = float(-9999)
outdir="F:\\DB_test_data\\VAR4\\"
for a,b,c,d,e,f,g in zip (CLDMASK ,NDVI ,BAND1 ,BAND2 ,BAND3 ,BAND4,AOD):
print ("processing:"+ a)
arcpy.AddMessage("processing:{}".format(b.split('\\')[4][9:30]))
name = c.split("\\")
filename=name[6][9:33]
print filename
arcpy.AddMessage("processing:{}".format(d.split('\\')[6][9:33]))
arcpy.AddMessage("processing:{}".format(e.split('\\')[6][9:30]))
arcpy.AddMessage("processing:{}".format(f.split('\\')[6][9:33]))
print ("processing:"+ g)
#######################################################################
setnull =arcpy.gp.SetNull_sa(g,g, "in_memory/dat", "\"Value\" = -9999")
ras=arcpy.Raster(setnull)
Data=(ras-add_offset)*Scale_factor
da = Con((Data >= 0.0) & (Data <= 0.1),1)
# Process: Extract by Cloud_Mask
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = "MAXOF"
ndvi_msk=arcpy.gp.ExtractByMask_sa(b, a)
Band1_mask=arcpy.gp.ExtractByMask_sa(c, a)
Band2_mask=arcpy.gp.ExtractByMask_sa(d, a)
Band3_mask=arcpy.gp.ExtractByMask_sa(e, a)
Band4_mask=arcpy.gp.ExtractByMask_sa(f, a)
arcpy.env.cellSize = tempEnvironment0
# Process: Extract by Mask using AOD less than 0.1 value
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = cellsize
ndvi_msk1=arcpy.gp.ExtractByMask_sa(ndvi_msk, da)
Band1_mask1=arcpy.gp.ExtractByMask_sa(Band1_mask, da)
Band2_mask2=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
Band3_mask3=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
Band4_mask4=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
arcpy.env.cellSize = tempEnvironment0
try:
# Raster to point
field ="VALUE"
Point=arcpy.RasterToPoint_conversion(ndvi_msk1,"in_memory/fileroot",field)
shape=arcpy.sa.ExtractMultiValuesToPoints(Point,[[ndvi_msk1,"NDVI"],[Band1_mask1,"Band1"],[Band2_mask2,"Band2"],
[Band3_mask3 ,"Band3"],[Band4_mask4,"Band4"]], "NONE")
#Add Two Fields Name and Name2
[arcpy.AddField_management(shape,field_name, "TEXT", field_length = 50)
for field_name in ["Name", "Name2"]]
arcpy.AddMessage("Successfully Added NAME fields ")
fieldss = ['Band1','Name']
with arcpy.da.UpdateCursor(shape, fieldss) as cursor:
for row in cursor:
if (row[0] > 0.02 and row[0] < 0.06):
row[1] = 'AAA'
print ("processing1:""{0:.8f}, {1}".format(row[0], row[1]))
elif (row[0] >= 0.06 and row[0] < 0.09):
row[1] = 'BBB'
# print ("processing1:""{0:.8f}, {1}".format(row[0], row[1]))
elif (row[0] >= 0.09):
row[1] = 'CCC'
else:
row[1] = 'NA'
# Update the cursor with the updated list
cursor.updateRow(row)
# Process: Select Layer By Attribute for separate the catagories "AAA","BBB,"CCC"
arcpy.MakeFeatureLayer_management(shape,"lyr1")
AAA = arcpy.SelectLayerByAttribute_management("lyr1", "NEW_SELECTION", "\"Name\" = 'AAA'")
print "Successfully Process : Select Layer By Attribute for AAA "
arcpy.MakeFeatureLayer_management(shape,"lyr2")
BBB = arcpy.SelectLayerByAttribute_management("lyr2", "NEW_SELECTION", "\"Name\" = 'BBB'")
print "Successfully Process : Select Layer By Attribute for BBB "
arcpy.MakeFeatureLayer_management(shape,"lyr3")
CCC = arcpy.SelectLayerByAttribute_management("lyr3", "NEW_SELECTION", "\"Name\" = 'CCC'")
print "Successfully Process : Select Layer By Attribute for CCC "
# Writing tables for categories("AAA","BBB,"CCC") and individual category have three Land type
# that will be save in different folder and their individual name
#------------------First we write category "AAA" and their indiviual land type--------------------------------------#
category="AAA"
file_name=filename
base_path = r"F:\\DB_test_data\\VAR4\\"
#def TableTotext(Input_Data,base_path,category,file_name):
#def Land_10():
land_use='LAND_10'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
else:
print ("Directory already exists" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['NDVI', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 10")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(AAA, fields) as cursor:
for row in cursor:
if (row[0] > 0.10 and row[0] < 0.20):
row[5] = 'landuse_10'
print ("processing_A:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write("{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 10 " )
#return Land_10
#def Land_20():
land_use='LAND_20'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
else:
print ("Directory already exists" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['NDVI', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 20")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(AAA, fields) as cursor:
for row in cursor:
if (row[0] >= 0.20 and row[0] < 0.50):
row[5] = 'landuse_20'
print ("processing_B:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write("{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 20 " )
#return Land_20
#def Land_30():
land_use='LAND_30'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
else:
print ("Directory already exists" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['NDVI', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 30")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(AAA, fields) as cursor:
for row in cursor:
if (row[0] >= 0.50):
row[5] = 'landuse_30'
print ("processing_C:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write("{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 30 " )
#return LAND_30
#------------------Second we write category "BBB" and their indiviual land type--------------------------------------#
print "Code is OK"
except:
print "Error in Code"
print arcpy.GetMessages()
print 'finished run: %s\n\n' % (datetime.datetime.now() - start)
Solved! Go to Solution.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Thank you Joshua Bixby Sir for sharing your views to clear the concept of Multiprocessing and single processing Function during the analysis of intensive data.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
I havent fully tested this, but heres a script that should hopefully show you how you could paramatise your "Tolerances", so you dont need to recode slightly different functions.
from operator import * #We can use operators now # gt = > # ge = >= #lt = < #le = <= minTolerance1= 0.10 minType1 = gt #Greater than maxTolerance1 = 0.20 maxType1 = lt #Less than minTolerance2 = 0.2 minType2 = ge #Greater Equal maxTolerance2 = 0.5 maxType2 = lt #Less than minTolerance3 = 0.5 minType3 = ge #Greater Equal maxTolerance3 = None maxType3 = None row = [0.2] def make_comparisons(row, minValue, minCheckType, maxValue, maxCheckType): if minCheckType == None and maxCheckType == None: #We have no comparison! result = "No comparison to make" elif minCheckType == None: #query with just max being checked result = maxCheckType(row[0], maxValue) elif maxCheckType == None: #Query just checking minimums result = minCheckType(row[0], minValue) else: result1 = maxCheckType(row[0], maxValue) result2 = minCheckType(row[0], minValue) if result1 == True and result2 == True: result = True else: result = False return result print make_comparisons(row, minTolerance1, minType1, maxTolerance1, maxType1) print make_comparisons(row, minTolerance2, minType2, maxTolerance2, maxType2) print make_comparisons(row, minTolerance3, minType3, maxTolerance3, maxType3)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Thank you, Luke Webb sir. I will try to manipulate my code as per your instruction. I think it will work fine.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Luke Webb sir, I just test your function and it works well in but the arguments
minTolerance3 = 0.5
minType3 = ge #Greater Equal
maxTolerance3 = None
maxType3 = Noneelif minCheckType == None: #query with just max being checked
result = maxCheckType(row[0], maxValue)
elif maxCheckType == None: #Query just checking minimums
result = minCheckType(row[0], minValue)
it does not work as my requirement (I need row[0] => 0.50 ) but it gives all < 0.5 > value
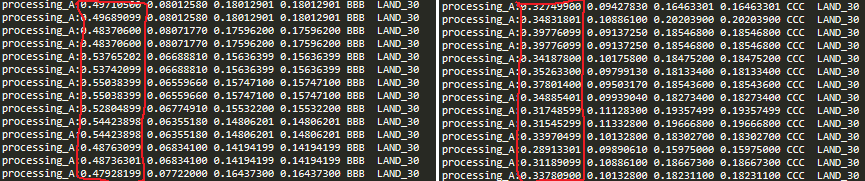
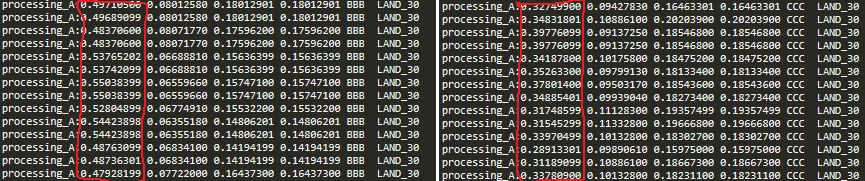

and my code is
def geoprocess (file_name,Input_Data,base_path,category,land_use,minValue, minCheckType, maxValue, maxCheckType):
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
try:
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
except OSError:
print ("Creation of the directory failed" )
else:
print ("Successfully created the directory" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['grid_code', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For:{0}".format(land_use))
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(Input_Data, fields) as cursor:
for row in cursor:
if minCheckType == None and maxCheckType == None: #We have no comparison!
result = "No comparison to make"
elif maxCheckType == None: #Query just checking minimums
result = minCheckType(row[0], minValue)
row[5] = land_use
print ("processing_A:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
elif minCheckType == None: #query with just max being checked
result = maxCheckType(row[0], maxValue)
row[5] = land_use
print ("processing_B:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
else:
result1 = minCheckType(row[0], minValue)
result2 = maxCheckType(row[0], maxValue)
if result1 == True and result2 == True:
row[5] = land_use
print ("processing_C:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
else:
result1 = False
txtfile.close()
print ("Successfully save the:{0}".format(land_use) )
print "Code is OK"
except:
print "Error in code"
def main(file_name,Input_Data,base_path,category):
minTolerance1= 0.10
minType1 = gt #Greater than
maxTolerance1 = 0.20
maxType1 = lt #Less than
minTolerance2 = 0.2
minType2 = ge #Greater Equal
maxTolerance2 = 0.5
maxType2 = lt #Less than
minTolerance3 = 0.5
minType3 = ge #Greater Equal
maxTolerance3 = None
maxType3 = None
geoprocess(file_name,Input_Data,base_path,category,'LAND_10',minTolerance1, minType1,maxTolerance1, maxType1)
geoprocess(file_name,Input_Data,base_path,category, 'LAND_20',minTolerance2, minType2, maxTolerance2, maxType2)
geoprocess(file_name,Input_Data,base_path,category, 'LAND_30', minTolerance3, minType3, maxTolerance3, maxType3)
try:
if __name__ == '__main__':
t1=time.time()
main(file_name,AAA,base_path,"OOO")
main(file_name,BBB,base_path,"FFF")
main(file_name,CCC,base_path,"ddd")
print "serial processing took:",time.time()-t1
print "serial processing is OK"
except:
print "Error in serial processing"
print arcpy.GetMessages()
print 'finished run: %s\n\n' % (datetime.datetime.now() - start)
please check this issue.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Luke Webb sir, are you checking my problem? I am waiting for your feedback.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
I looked into it without conclusion my code seems good, this returns the correct values: FALSE, TRUE, TRUE
from operator import * minTolerance = 0.5 minType = ge #Greater Equal maxTolerance = None maxType = None row1 = [0.4] row2 = [0.5] row3 = [0.6] def make_comparisons(row, minValue, minCheckType, maxValue, maxCheckType): if minCheckType == None and maxCheckType == None: #We have no comparison! result = "No comparison to make" elif minCheckType == None: #query with just max being checked result = maxCheckType(row[0], maxValue) elif maxCheckType == None: #Query just checking minimums result = minCheckType(row[0], minValue) else: result1 = maxCheckType(row[0], maxValue) result2 = minCheckType(row[0], minValue) if result1 == True and result2 == True: result = True else: result = False return result print make_comparisons(row1, minTolerance, minType, maxTolerance, maxType) print make_comparisons(row2, minTolerance, minType, maxTolerance, maxType) print make_comparisons(row3, minTolerance, minType, maxTolerance, maxType)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Luke Webb sir, Have there any problem within my code which I wrote earlier?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
I did notice the issue, you are not actually using the result of the comparison.
For example instead of:
result = minCheckType(row[0], minValue)
row[5] = land_use
print ("processing_A:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
You should have done:
result = minCheckType(row[0], minValue)
if result == True:
row[5] = land_use
print ("processing_A:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
Sadly did not spot this issue at first and am hopelessly addicted to python, so I rewrote your script here to show you how to use functions a bit more, there may be a bug as i didnt test it ![]() :
:
__author__ = 'Beast2' import os from operator import * import time import datetime import arcpy import sys def create_output_directory(outdir): if not os.path.exists(outdir): try: print('\nCreating new output directory!\n'+outdir+'\n') os.makedirs(outdir) except OSError: print ("Creation of the directory failed" ) else: print ("Directory Already Exists") return True def make_comparisons(value, minCheckType, minValue, maxCheckType, maxValue): if minCheckType == None and maxCheckType == None: #We have no comparison! result = "No comparison to make" elif minCheckType == None: #query with just max being checked result = maxCheckType(value, maxValue) elif maxCheckType == None: #Query just checking minimums result = minCheckType(value, minValue) else: result1 = maxCheckType(value, maxValue) result2 = minCheckType(value, minValue) if result1 == True and result2 == True: result = True else: result = False return result def main(file_name, base_path, categories, land_uses): print "Processing to file name: %s" % file_name print "Saving results to: %s" % base_path print "Processing %s categories" % len(categories) for category in categories: print "Processing Category: %s" % category[0] print "Category Input Data: %s" % category[1] for land_use in land_uses: output_directory_path = os.path.join(base_path, category[0], land_use[0]) print "Processing Land Use: %s" % land_use[0] create_output_directory(output_directory_path) ras_name = os.path.join(output_directory_path,("{0}_{1}{2}_{3}".format(category[0],file_name,land_use[0],".txt"))) fields = ['grid_code', 'Band1', 'Band2','Band3','Name',"Name2"] with open(ras_name, 'wb') as txtfile: arcpy.AddMessage("fields Writing For:{0}".format(land_use[0])) txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5])) with arcpy.da.UpdateCursor(category[1], fields) as cursor: for row in cursor: row_test_value = row[0] #Check if the row value, is within the land use category if make_comparisons(row_test_value, land_use[1], land_use[2], land_use[3], land_use[4]) == True: row[5] = land_use[0] print ("processing:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5])) txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5])) txtfile.close() print ("Successfully save the:{0}".format(land_use[0])) if __name__ == '__main__': try: start_time = time.time() #Categories we will process category_1 = ['OOO', 'input_data'] category_2 = ['FFF', 'input_data'] category_3 = ['ddd', 'input_data'] categories = [category_1, category_2, category_3] #Land Use category information land_10 = ['LAND_10', gt, 0.1, lt, 0.2] land_20 = ['LAND_20', ge, 0.2, lt, 0.5] land_30 = ['LAND_30', ge, 0.5, None, None] land_uses = [land_10, land_20, land_30] #Other parameters base_path = 'base_path' file_name = 'file_name' main(file_name, base_path, categories, land_uses) print "serial processing took:",time.time() - start_time print "serial processing is OK" except: print "Error in serial processing" print arcpy.GetMessages() print 'finished run: %s\n\n' % (datetime.datetime.now() - start_time)py
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Thank you Luke Webb sir, your code working well as per my requirement. And also thank full to other persons who continuously helped me.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Glad to hear!
Now, I know nothing about multiprocessing, but I think by just making a small change, copying the "process" code into its own function, we could now implement multiprocessing easily, as the main script will build the list of processes to be run. Something like this:
__author__ = 'Beast2'
import os
from operator import *
import time
import datetime
import arcpy
import sys
def create_output_directory(outdir):
if not os.path.exists(outdir):
try:
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
except OSError:
print ("Creation of the directory failed" )
else:
print ("Directory Already Exists")
return True
def make_comparisons(value, minCheckType, minValue, maxCheckType, maxValue):
if minCheckType == None and maxCheckType == None: #We have no comparison!
result = "No comparison to make"
elif minCheckType == None: #query with just max being checked
result = maxCheckType(value, maxValue)
elif maxCheckType == None: #Query just checking minimums
result = minCheckType(value, minValue)
else:
result1 = maxCheckType(value, maxValue)
result2 = minCheckType(value, minValue)
if result1 == True and result2 == True:
result = True
else:
result = False
return result
def geoprocess(file_name, base_path, category, land_use):
output_directory_path = os.path.join(base_path, category[0], land_use[0])
print "Processing Land Use: %s" % land_use[0]
create_output_directory(output_directory_path)
ras_name = os.path.join(output_directory_path,("{0}_{1}{2}_{3}".format(category[0],file_name,land_use[0],".txt")))
fields = ['grid_code', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For:{0}".format(land_use[0]))
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(category[1], fields) as cursor:
for row in cursor:
row_test_value = row[0]
#Check if the row value, is within the land use category
if make_comparisons(row_test_value, land_use[1], land_use[2], land_use[3], land_use[4]) == True:
row[5] = land_use[0]
print ("processing:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the:{0}".format(land_use[0]))
def main(file_name, base_path, categories, land_uses):
print "Processing to file name: %s" % file_name
print "Saving results to: %s" % base_path
print "Processing %s categories" % len(categories)
for category in categories:
print "Processing Category: %s" % category[0]
print "Category Input Data: %s" % category[1]
for land_use in land_uses:
#Implement Multi Processing Here to run all X processes at same time
a_process_that_could_run_parrallel_to_others = geoprocess(file_name, base_path, category, land_use)
if __name__ == '__main__':
try:
start_time = time.time()
#Categories we will process
category_1 = ['OOO', 'input_data']
category_2 = ['FFF', 'input_data']
category_3 = ['ddd', 'input_data']
categories = [category_1, category_2, category_3]
#Land Use category information
land_10 = ['LAND_10', gt, 0.1, lt, 0.2]
land_20 = ['LAND_20', ge, 0.2, lt, 0.5]
land_30 = ['LAND_30', ge, 0.5, None, None]
land_uses = [land_10, land_20, land_30]
#Other parameters
base_path = 'base_path'
file_name = 'file_name'
main(file_name, base_path, categories, land_uses)
print "serial processing took:",time.time() - start_time
print "serial processing is OK"
except:
print "Error in serial processing"
print arcpy.GetMessages()
print 'finished run: %s\n\n' % (datetime.datetime.now() - start_time)