It seemed like a simple idea at the time.
Take the outputs from another data source, concatenate the values to a string and toss them into a new field.
Looks nice eh?
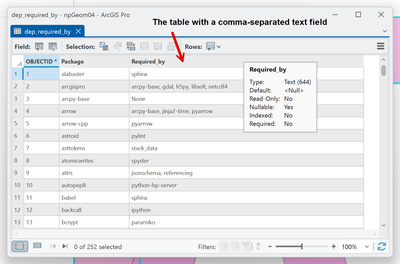
Well, well, you can even query the whole stringy-thing. Don't get me wrong, the whole Select By Attributes and the sql thing are pretty cool
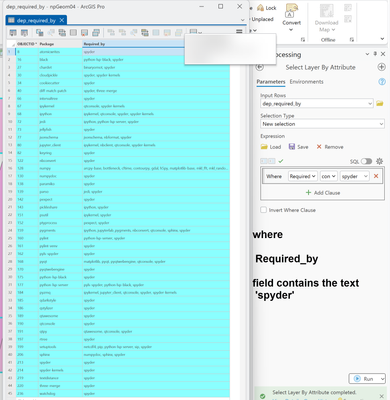
Sorting is fun too. Who would have thought that arcpy-base had so many friends (dependencies).
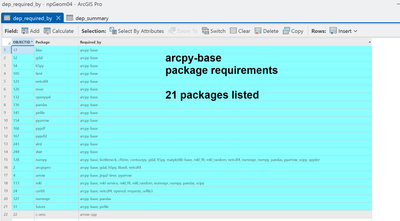
That is when I asked myself what are the interdependencies/requirements/dependencies for Spyder, my favorite python IDE. Sql away, the sort thing got me to roll.
That is when I thought .... what about this package? how about that one?
Time for a big summary, which wasn't going to work with the existing options.
That is when I remembered those arc angels that coded the numpy stuff!! (not the post stevie ray band or things divinical)
TableToNumPyArray and NumPyArrayToTable.
Follow along.
1 import arcpy of course or
2-4 specify your table of interest and its field(s). We now have an array. None of this "for row in cursor stuff".
'vals' is a view of the data in 'arr', so we can explore it and manipulate it
6 list comprehensions can be fun
- for every value (v) in the array (vals)
- split the value at the commas (note I used "," instead of ", " since there was a mixture
- strip off any leading/trailing spaces to be on the safe side
7 flatten the whole dataset
8 get the unique entries and their counts. Remember the field was one big messy string/former list thing.
9- 11 Create the output array and send the whole summary back to Pro
import arcpy
tbl = r"C:\arcpro_npg\Project_npg\npgeom.gdb\dep_required_by"
arr = arcpy.da.TableToNumPyArray(tbl, "Required_by")
vals = arr["Required_by"]
big = [i.strip() for v in vals for i in v.split(",")]
big_flat = npg.flatten(big)
uniq1, cnts1 = np.unique(big_flat, return_counts=True)
out_tbl = r"C:\arcpro_npg\Project_npg\npgeom.gdb\dep_summary"
out_arr = np.asarray(list(zip(uniq1, cnts1)), dtype=[('Package', 'U50'), ('Counts', 'i4')])
arcpy.da.NumPyArrayToTable(out_arr, out_tbl)
Now the next time I am interested in exploring the python package infrastructure for Pro, I have a workflow.
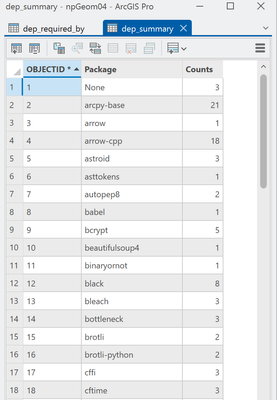
Don't be afraid to use what was given to you. Arcpy and NumPy play nice together.