- Home
- :
- All Communities
- :
- Products
- :
- ArcGIS Pro
- :
- ArcGIS Pro Questions
- :
- Re: Calculate field/ python code question
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Good day all,
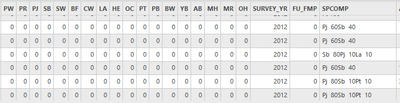
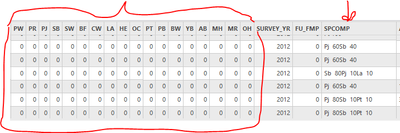
I have a layer with the following fields in the attribute table:
I am trying to populate the species fields (PW, PR, PJ, SB, etc.) with the information found in the "SPCOMP" field.
All of the values that are in the “SPCOMP” field should be put into related individual species fields, and leave the rest of species fields to 0. For example, if the “SPCOMP” = “Pj 60Pt 30Bw 10”, that means that Pj=60, Pt=30, Bw=10. Then 60 should go under “PJ”, 30 goes under “PT”, and 10 goes under “BW”.
I've tried various different things through the "Calculate field" tool but have been unsuccessful.
I am thinking something like:
For field PW: if SPCOMP includes the text "Pw" return following number - omit the rest of the string, else return 0.
Obviously this is not in proper Python code, and would have to be repeated for every field.
If anyone has a solution, or Python code example to accomplish this, I would be so grateful.
Thank you
Solved! Go to Solution.
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Although a regular expression answer has been put forward, I believe there is a more straightforward regular expression than what has been already proposed. Also, all that is needed in terms of cursors is a single pass through the data set with an update cursor.
First, the regular expression. You are trying to match pairs of species-values that have been concatenated into a single string, so there are 3 things to differentiate: 1) a species-value pair, 2) the species within that pair, and 3) the numeric value for the species in that pair.
The following regular expression captures that logic:
>>> import re
>>>
>>> reg_exp = "(?:([A-Za-z]+)\s*([0-9]+))+?"
>>>
>>> SPCOMP_samples = [
... "Pj 60Sb 40",
... "Pj 60Sb 40",
... "Sb 80Pj 10La 10",
... "Pj 60Sb 40",
... "Pj 80Sb 10Pt 10",
... "Pj 80Sb 10Pt 10"
... ]
>>>
>>> for sample in SPCOMP_samples:
... re.findall(reg_exp, sample)
...
[('Pj', '60'), ('Sb', '40')]
[('Pj', '60'), ('Sb', '40')]
[('Sb', '80'), ('Pj', '10'), ('La', '10')]
[('Pj', '60'), ('Sb', '40')]
[('Pj', '80'), ('Sb', '10'), ('Pt', '10')]
[('Pj', '80'), ('Sb', '10'), ('Pt', '10')]
>>>
The regular expression uses a non-capturing outer group to find the species-value pairs, and then uses two internal capturing groups to differentiate the species from the numeric value. Using the expression with re.findall returns a list of tuples containing a species and a numeric value.
Since each tuple contains the species and the value, you can use the species to look up the index of the species field in a cursor and update that field with the value.
# Note: Code below hasn't been tested
reg_exp = "(?:([A-Za-z]+)\s*([0-9]+))+?"
fc = # path to feature class or shape file
with arcpy.da.UpdateCursor(fc, "*") as cur:
spcomp_idx = cur.fields.index("SPCOMP")
for row in cur:
for species, value in re.findall(reg_exp, row[spcomp_idx]):
species_idx = cur.fields.index(species.upper())
row[species_idx] = value
cur.updateRow(row)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Take that offending line out entirely.
In addition to the indent error, that line is redefining the fc variable to be ???? because you haven't put a value there, just a comment telling you to put a value there. (Edit: this will throw an end of line error, since it's expecting a value for fc. It just hit the indent error first.) You already defined the fc variable at the top, so you don't need it here. (Probably the same for the second instance of the regular expression)
Python relies on indentation to figure out how each line relates to each other, lines belonging to a loop are indented further than the loop open itself.
e.g.
for i in [1,2,3]:
#Do something
# Do something else
In this case, the offending line has one space at the beginning, throwing it out of line with everything else.
Also, so does the following line (with arcpy.da.UpdateCursor...)
Generally, 4 spaces (Or a tab, pick one or the other but not both) is recommended per indent level, but I think technically as long as they're consistent within whatever loop they belong to, you're fine. Four spaces is better for readability, though.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
your spcomp field is poorly combined
PJ60 PT30 BW10
would have allowed one to split the inputs on the spaces.
Are you stuck with that column? If so, are all the numbers in increments of 10? (eg 10 20 30.... 90 (no 0 or 100)
... sort of retired...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hello Dan,
Since this was an exercise I was given to test my coding skills (which are very limited - I have only used python a handful of times, for very basic stuff) - if reorganizing the column and keeping the data intact allows for an easier solution to my problem - then I think this is a good way to go.
I also see what you are saying with regards to its formatting "Pj 10Sb 60 La 30" should infact be written: "Pj10 Sb60 La30".
Although, rearranging the column format would necessitate more python code I am not familiar with...
The numbers in the SPCOMP field are all in increments of 10 - ranging from 10 to 100 (10 % to 100% species composition).
I really appreciate the comment and ideas to get me on the right path.
Thank you.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
There's always regex if you're brave.
This assumes that your fields are always going to match the species codes. It's also more or less untested.
The basic workflow is:
- Cycle through the table and create dictionary of objectid: {species: number} using regex to find all instances of two letters followed by one or more spaces and any number of digits in the SPCOMP field.
- Cycle through again with an update cursor and update the rows as needed.
You could probably consolidate this all into a single update cursor, but I didn't have time today to think it through.
Anyway, I think (hope) that this is at least somewhat useful.
import re
fc = #your feature class
specDict ={}
fieldList = ["OBJECTID"]
# Search cursor, split the SPCOMP and load into dictionary.
# Populate field list to calculate fields later.
with arcpy.da.SearchCursor(fc, ['OBJECTID', 'SPCOMP']) as cursor:
for row in cursor:
# Find all instances of two letters, followed by one or more spaces and
# any number of numbers.
for line in re.findall(r"([a-zA-z]{2} +\d*)", row[1]):
# You could probably do .split()[0] and [1] here but
# I'm on a roll with the regex.
num = re.match(r"\d", line)
sp = re.match(r"[a-zA-z]{2}", line)
# num = line.split(" ")[1]
# sp = line.split(" ")[0]
# Add species to fieldList
# You could probably comment this out and manually populate
# the fieldList
if sp not in fieldList:
fieldList.append(sp)
# Add to the dictionary.
# specDict = {ObjectID: {Species 1: Number, Species 2: Number...}}
if str(row[0]) in specDict:
specDict[str(row[0])][sp] = num
else:
specDict[str(row[0])] = {sp: num}
with arcpy.da.UpdateCursor(fc, fieldList) as cursor:
for row in cursor:
# Search through the field list and get the index of the field
# in the row tuple, then apply the the correct value for that
# species to that field.
for field in fieldList:
if field in specDict[str(row[0])]:
row[row.index(field)] = specDict[row[0]][field]
cursor.updateRow(row)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hello Alfred,
I greatly appreciate you taking the time to type all this out for me.
I am fairly new to python and have only used it a handful of times to perform basic tasks. Not at all familiar with regex, unfortunately.
I will try the code you provided and see if I am able to perform the required task.
Thank you.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
No worries, I just tested it and found some issues. Will upload an update shortly.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Sorry about that. The basis of the code was correct, but I forgot how to work with regex a bit (I only recently began to understand how it works) and had to change things around. (Also this code assumes that your species fields are integers. If they're not, you might have to play around and convert to text or something. Similarly, it assumes that each species field matches the value in SPCOMP exactly.)
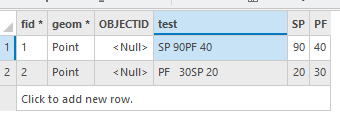
My fields are a bit different (so I used fid and test instead of OBJECTID and SPCOMP), but below is the before and after. I made sure to test with any number of spaces.
One final thing: If you run an update cursor with the attribute table open, you have to refresh the table to see the changes. Also, this was written for a file gdb. If you're using an enterprise gdb, it gets a little bit more complicated.
import re #Edited to add this line in since I forgot it whoops.
fc = r"Trail_MDB1.gdb\testingFC"
specDict ={}
fieldList = ['OBJECTID']
# Search cursor, split the SPCOMP and load into dictionary.
# Populate field list to calculate fields later.
with arcpy.da.SearchCursor(fc, ['OBJECTID', 'SPCOMP']) as cursor:
for row in cursor:
# Find all instances of two letters, followed by one or more spaces and
# any number of numbers.
for line in re.findall(r"([a-zA-z]{2} +\d+)", row[1]):
# You could probably do .split()[0] and [1] here but
# I'm on a roll with the regex.
sp = re.match(r"[a-zA-z]{2}", line).group(0)
#I have no idea why, but the match doesn't like the number
# when there are also letters in the string
# so we're using search instead.
num = re.search(r"(\d+)", line).group(0)
# num = line.split(" ")[1]
# sp = line.split(" ")[0]
# Add species to fieldList
# You could probably comment this out and manually populate
# the fieldList
if sp not in fieldList:
fieldList.append(sp)
# Add to the dictionary.
# specDict = {ObjectID: {Species 1: Number, Species 2: Number...}}
if str(row[0]) in specDict:
specDict[str(row[0])][sp] = num
else:
specDict[str(row[0])] = {sp: num}
with arcpy.da.UpdateCursor(fc, fieldList) as cursor:
for row in cursor:
# Search through the field list and get the index of the field
# in the row tuple, then apply the the correct value for that
# species to that field.
for field in fieldList:
if field in specDict[str(row[0])]:
row[fieldList.index(field)] = specDict[str(row[0])][field]
cursor.updateRow(row)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hello Alfred,
Again, I really appreciate your time and effort in trying to find this solution.
As I am trying to perform this task with a shapefile - the above code is no working.
I will keep trying things though.
Thanks again.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi Brodie, just tested it on a shapefile and it worked fine.
If it's not giving you an error message, then I assume you just have to refresh the table for it to show up correctly.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
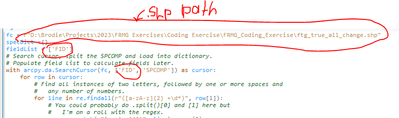
Hi Alfred,
I confirmed that my SPCOMP field is in fact text format.
Made the following changes to the code provided:
I changed OBJECTID to FID since I did not have an OBJECTID field:
Here is the error I am receiving:
Line 10 being:
for line in re.findall(r"([a-zA-z]{2} +\d*)", row[1]):
Again your help is invaluable.
Thank you.