- Home
- :
- All Communities
- :
- Developers
- :
- Python
- :
- Python Questions
- :
- Re: Write ArcPy cursor results to a text file in d...
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Write ArcPy cursor results to a text file in different folders with a different name based on cursor condition. Define function is not working.Multiprocessing
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Middle section of the code, I separate the attribute value in a different category ("AAA","BBB","CCC") and every category have Ten land-use types, initially, I set Three land use type (LAND_10, LAND_20, LAND_30) for test code. Now I used the update cursor for different land-use type(LAND_10, LAND_20, LAND_30) , and saved the text file to there corresponding Land use folders(Like, Land_10 ....Land_30) under the "AAA" folder. similarly, this function I will use for categories "BBB","CCC" .
Initially, I write this code for category "AAA" but I want to use this function for category "BBB","CCC" by defining a function(uncheck all define function), but is not working. Below is my code please check it.
First of all, I want to reduce my code (Because I have 10 land type initially I write three) and the whole function I want to use as a defined function for a different category.
Another thing is Multiprocessing, Multithreading I want to use this function for the code because of initially my code takes much more time for one processes. In my case how to use the multiprocessing function.
please look at this matter and give me a suggestion to manipulate this code.
import datetime
start = datetime.datetime.now()
print 'start run: %s\n' % (start)
import arcpy ,os ,sys,csv,errno
from arcpy import env
from arcpy.sa import *
import datetime
import re
import glob
import itertools
arcpy.env.overwriteOutput = True
import multiprocessing as mp
print("Number of processors: ", mp.cpu_count())
cellsize = "F:\\DB_test_data\\TEST_RAY\\TEST1\\MOD02HKM_A2017001_0530_NDVI_AA.img"
d1="F:\\DB_test_data\\VAR2\\cldmask\\tt"
CLDMASK = glob.glob(d1 + os.sep + "*.Aerosol_Cldmask_Land_Ocean-Aerosol_Cldmask_Land_Ocean.tif")
CLDMASK.sort()
if CLDMASK is None:
print 'Could not open the CLDMASK raster files'
sys.exit(1)
else:
print 'The CLDMASK raster files was opened successfully'
#print CLDMASK
d2="F:\\DB_test_data\\TEST_RAY\\TEST1"
NDVI = glob.glob(d2 + os.sep + "*A2017001_0530_NDVI_AA.img")
NDVI.sort()
if NDVI is None:
print 'Could not open the NDVI raster files'
sys.exit(1)
else:
print 'The NDVI raster files was opened successfully'
#print NDVI
d3="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND1 = glob.glob(d3 + os.sep + "*A2017001_0530_006_BAND_1.img")
BAND1.sort()
if BAND1 is None:
print 'Could not open the BAND1 raster files'
sys.exit(1)
else:
print 'The BAND1 raster files was opened successfully'
#print BAND1
d4="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND2 = glob.glob(d4 + os.sep + "*A2017001_0530_006_BAND_2.img")
BAND2.sort()
if BAND2 is None:
print 'Could not open the BAND2 raster files'
sys.exit(1)
else:
print 'The BAND2 raster files was opened successfully'
#print BAND2
d4="F:\\DB_test_data\\VAR2\\IDL\\MOD09\\TEST"
BAND3 = glob.glob(d4 + os.sep + "*A2017001_0530_006_BAND_3.img")
BAND3.sort()
if BAND3 is None:
print 'Could not open the BAND3 raster files'
sys.exit(1)
else:
print 'The BAND3 raster files was opened successfully'
#print BAND3
d5="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND4 = glob.glob(d5 + os.sep + "*A2017001_0530_006_BAND_4.img")
BAND4.sort()
if BAND4 is None:
print 'Could not open the BAND4 raster files'
sys.exit(1)
else:
print 'The BAND4 raster files was opened successfully'
#print BAND4
d6 = r"F:\\DB_test_data\\VAR2\\MOD04_l2"
AOD = glob.glob(d6 + os.sep + "*Corrected_Optical_Depth_Land_2-Corrected_Optical_Depth_Land.tif")
AOD.sort()
if AOD is None:
print 'Could not open the AOD raster files'
sys.exit(1)
else:
print 'The AOD raster files was opened successfully'
Scale_factor = float(0.0010000000474974513)
add_offset = float(0.0)
Fill_value = float(-9999)
outdir="F:\\DB_test_data\\VAR4\\"
for a,b,c,d,e,f,g in zip (CLDMASK ,NDVI ,BAND1 ,BAND2 ,BAND3 ,BAND4,AOD):
print ("processing:"+ a)
arcpy.AddMessage("processing:{}".format(b.split('\\')[4][9:30]))
name = c.split("\\")
filename=name[6][9:33]
print filename
arcpy.AddMessage("processing:{}".format(d.split('\\')[6][9:33]))
arcpy.AddMessage("processing:{}".format(e.split('\\')[6][9:30]))
arcpy.AddMessage("processing:{}".format(f.split('\\')[6][9:33]))
print ("processing:"+ g)
#######################################################################
setnull =arcpy.gp.SetNull_sa(g,g, "in_memory/dat", "\"Value\" = -9999")
ras=arcpy.Raster(setnull)
Data=(ras-add_offset)*Scale_factor
da = Con((Data >= 0.0) & (Data <= 0.1),1)
# Process: Extract by Cloud_Mask
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = "MAXOF"
ndvi_msk=arcpy.gp.ExtractByMask_sa(b, a)
Band1_mask=arcpy.gp.ExtractByMask_sa(c, a)
Band2_mask=arcpy.gp.ExtractByMask_sa(d, a)
Band3_mask=arcpy.gp.ExtractByMask_sa(e, a)
Band4_mask=arcpy.gp.ExtractByMask_sa(f, a)
arcpy.env.cellSize = tempEnvironment0
# Process: Extract by Mask using AOD less than 0.1 value
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = cellsize
ndvi_msk1=arcpy.gp.ExtractByMask_sa(ndvi_msk, da)
Band1_mask1=arcpy.gp.ExtractByMask_sa(Band1_mask, da)
Band2_mask2=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
Band3_mask3=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
Band4_mask4=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
arcpy.env.cellSize = tempEnvironment0
try:
# Raster to point
field ="VALUE"
Point=arcpy.RasterToPoint_conversion(ndvi_msk1,"in_memory/fileroot",field)
shape=arcpy.sa.ExtractMultiValuesToPoints(Point,[[ndvi_msk1,"NDVI"],[Band1_mask1,"Band1"],[Band2_mask2,"Band2"],
[Band3_mask3 ,"Band3"],[Band4_mask4,"Band4"]], "NONE")
#Add Two Fields Name and Name2
[arcpy.AddField_management(shape,field_name, "TEXT", field_length = 50)
for field_name in ["Name", "Name2"]]
arcpy.AddMessage("Successfully Added NAME fields ")
fieldss = ['Band1','Name']
with arcpy.da.UpdateCursor(shape, fieldss) as cursor:
for row in cursor:
if (row[0] > 0.02 and row[0] < 0.06):
row[1] = 'AAA'
print ("processing1:""{0:.8f}, {1}".format(row[0], row[1]))
elif (row[0] >= 0.06 and row[0] < 0.09):
row[1] = 'BBB'
# print ("processing1:""{0:.8f}, {1}".format(row[0], row[1]))
elif (row[0] >= 0.09):
row[1] = 'CCC'
else:
row[1] = 'NA'
# Update the cursor with the updated list
cursor.updateRow(row)
# Process: Select Layer By Attribute for separate the catagories "AAA","BBB,"CCC"
arcpy.MakeFeatureLayer_management(shape,"lyr1")
AAA = arcpy.SelectLayerByAttribute_management("lyr1", "NEW_SELECTION", "\"Name\" = 'AAA'")
print "Successfully Process : Select Layer By Attribute for AAA "
arcpy.MakeFeatureLayer_management(shape,"lyr2")
BBB = arcpy.SelectLayerByAttribute_management("lyr2", "NEW_SELECTION", "\"Name\" = 'BBB'")
print "Successfully Process : Select Layer By Attribute for BBB "
arcpy.MakeFeatureLayer_management(shape,"lyr3")
CCC = arcpy.SelectLayerByAttribute_management("lyr3", "NEW_SELECTION", "\"Name\" = 'CCC'")
print "Successfully Process : Select Layer By Attribute for CCC "
# Writing tables for categories("AAA","BBB,"CCC") and individual category have three Land type
# that will be save in different folder and their individual name
#------------------First we write category "AAA" and their indiviual land type--------------------------------------#
category="AAA"
file_name=filename
base_path = r"F:\\DB_test_data\\VAR4\\"
#def TableTotext(Input_Data,base_path,category,file_name):
#def Land_10():
land_use='LAND_10'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
else:
print ("Directory already exists" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['NDVI', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 10")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(AAA, fields) as cursor:
for row in cursor:
if (row[0] > 0.10 and row[0] < 0.20):
row[5] = 'landuse_10'
print ("processing_A:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write("{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 10 " )
#return Land_10
#def Land_20():
land_use='LAND_20'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
else:
print ("Directory already exists" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['NDVI', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 20")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(AAA, fields) as cursor:
for row in cursor:
if (row[0] >= 0.20 and row[0] < 0.50):
row[5] = 'landuse_20'
print ("processing_B:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write("{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 20 " )
#return Land_20
#def Land_30():
land_use='LAND_30'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
else:
print ("Directory already exists" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['NDVI', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 30")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(AAA, fields) as cursor:
for row in cursor:
if (row[0] >= 0.50):
row[5] = 'landuse_30'
print ("processing_C:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write("{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 30 " )
#return LAND_30
#------------------Second we write category "BBB" and their indiviual land type--------------------------------------#
print "Code is OK"
except:
print "Error in Code"
print arcpy.GetMessages()
print 'finished run: %s\n\n' % (datetime.datetime.now() - start)
Solved! Go to Solution.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Thank you sir...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
What is your final suggestion for my code? Please, sir, refer me any who can resolve this matter.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
I'm pretty much tapped out for suggestions: my approach would be to modularize the code so it's easier to follow the logic as well as capture any errors encountered along the way.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
How do I configure my code with multiprocessing and multithreading?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Why the developer not focusing on my problem?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Base on my previous code I am trying to used multiprocessing function but is not working. please check this issue and give me a suggestion to resolve this issue. Joshua Bixby Xander BakkerDan Patterson
if __name__ == "__main__":
category="AAA"
file_name="MOD022"
base_path = r"F:\\DB_test_data\\VAR4\\"
try:
t1=time.time()
main(base_path,category,file_name,AAA)
print "serial processing took:",time.time()-t1
print "serial processing is OK"
except Exception as e:
print "Error in serial processing"
print(err)
try:
t2=time.time()
pool = multiprocessing.Pool(processes=4)
A=pool.map(main,(base_path,"AAA",file_name,AAA))
B=pool.map(main,(base_path,"BBB",file_name,BBB))
C=pool.map(main,(base_path,"CCC",file_name,CCC))
A.close()
B.close()
C.close()
A.join()
B.join()
C.join()
print "Pool took",time.time()-t2
print "multiprocessing is OK"
except Exception as e:
print "Error in multiprocessing "
#print(err)- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Skip multiprocessing for now and show what the inputs and outputs are supposed to look like. I am still seeing streams of code without knowing what the process is intending to do. I don't even know if multi or parallel processing is the route to go. There is a false assumption that this makes things better when indeed it is not necessarily the case.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
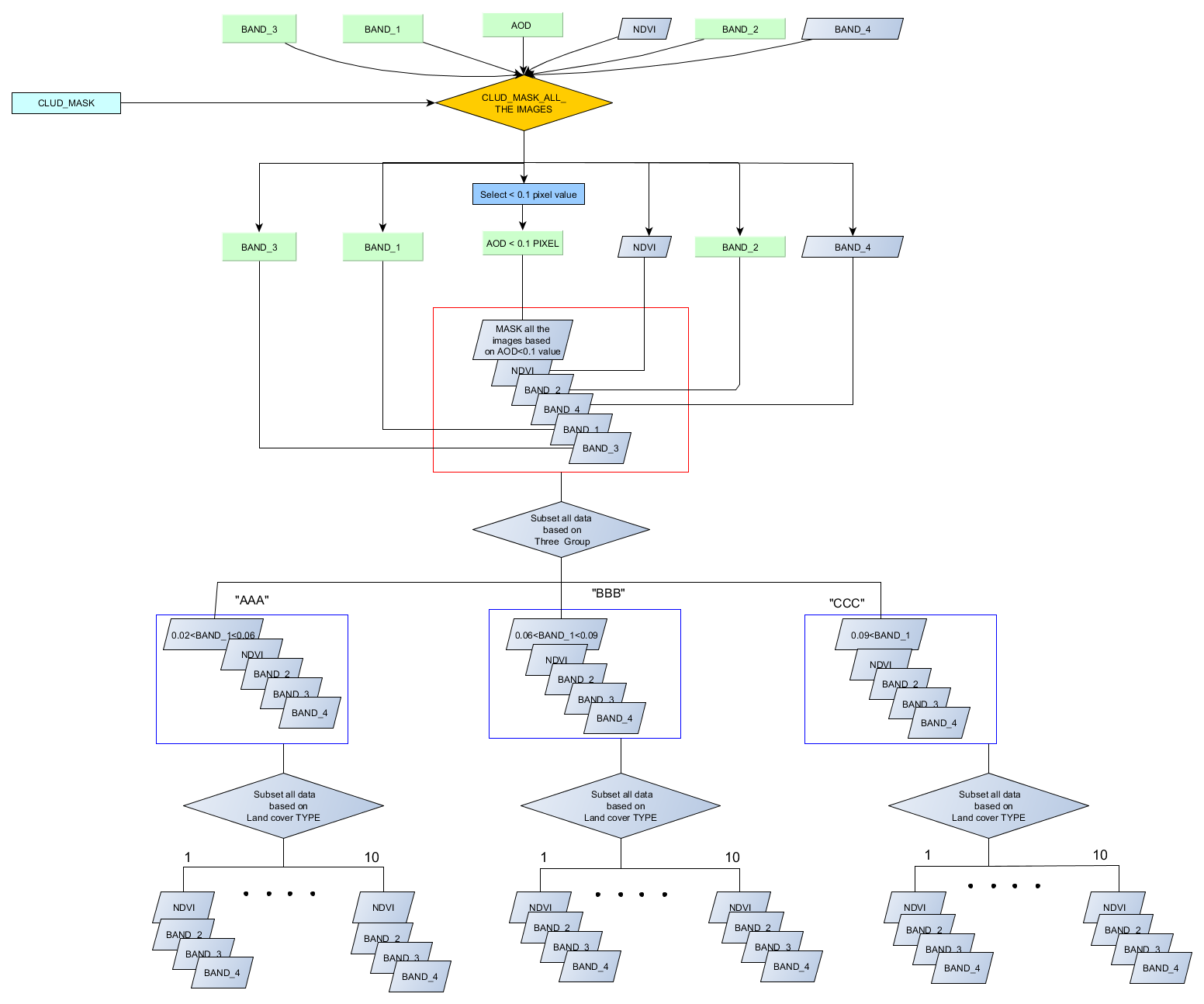
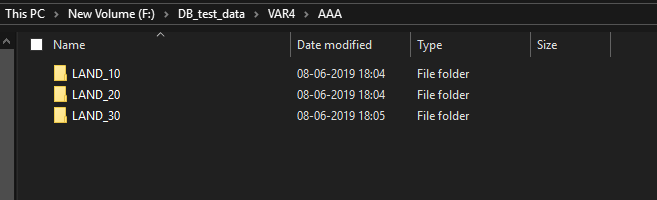
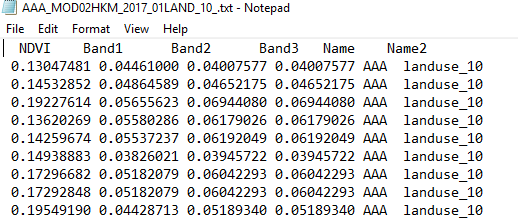
Here I attach my working flow and folder destination system and output files. I think you will understand what I want to do.
import datetime
start = datetime.datetime.now()
print 'start run: %s\n' % (start)
import arcpy ,os ,sys,csv,errno
from arcpy import env
from arcpy.sa import *
import datetime
import re
import glob
import itertools
arcpy.env.overwriteOutput = True
import multiprocessing
print("Number of processors: ", multiprocessing.cpu_count())
cellsize = "F:\\DB_test_data\\TEST_RAY\\TEST1\\MOD02HKM_A2017001_0530_NDVI_AA.img"
d1="F:\\DB_test_data\\VAR2\\cldmask\\tt"
CLDMASK = glob.glob(d1 + os.sep + "*.Aerosol_Cldmask_Land_Ocean-Aerosol_Cldmask_Land_Ocean.tif")
CLDMASK.sort()
if CLDMASK is None:
print 'Could not open the CLDMASK raster files'
sys.exit(1)
else:
print 'The CLDMASK raster files was opened successfully'
#print CLDMASK
d2="F:\\DB_test_data\\TEST_RAY\\TEST1"
NDVI = glob.glob(d2 + os.sep + "*A2017001_0530_NDVI_AA.img")
NDVI.sort()
if NDVI is None:
print 'Could not open the NDVI raster files'
sys.exit(1)
else:
print 'The NDVI raster files was opened successfully'
#print NDVI
d3="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND1 = glob.glob(d3 + os.sep + "*A2017001_0530_006_BAND_1.img")
BAND1.sort()
if BAND1 is None:
print 'Could not open the BAND1 raster files'
sys.exit(1)
else:
print 'The BAND1 raster files was opened successfully'
#print BAND1
d4="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND2 = glob.glob(d4 + os.sep + "*A2017001_0530_006_BAND_2.img")
BAND2.sort()
if BAND2 is None:
print 'Could not open the BAND2 raster files'
sys.exit(1)
else:
print 'The BAND2 raster files was opened successfully'
#print BAND2
d4="F:\\DB_test_data\\VAR2\\IDL\\MOD09\\TEST"
BAND3 = glob.glob(d4 + os.sep + "*A2017001_0530_006_BAND_3.img")
BAND3.sort()
if BAND3 is None:
print 'Could not open the BAND3 raster files'
sys.exit(1)
else:
print 'The BAND3 raster files was opened successfully'
#print BAND3
d5="F:\\DB_test_data\\VAR2\\IDL\\MOD02HKM\\TEST"
BAND4 = glob.glob(d5 + os.sep + "*A2017001_0530_006_BAND_4.img")
BAND4.sort()
if BAND4 is None:
print 'Could not open the BAND4 raster files'
sys.exit(1)
else:
print 'The BAND4 raster files was opened successfully'
#print BAND4
d6 = r"F:\\DB_test_data\\VAR2\\MOD04_l2"
AOD = glob.glob(d6 + os.sep + "*Corrected_Optical_Depth_Land_2-Corrected_Optical_Depth_Land.tif")
AOD.sort()
if AOD is None:
print 'Could not open the AOD raster files'
sys.exit(1)
else:
print 'The AOD raster files was opened successfully'
Scale_factor = float(0.0010000000474974513)
add_offset = float(0.0)
Fill_value = float(-9999)
outdir="F:\\DB_test_data\\VAR4\\"
for a,b,c,d,e,f,g in zip (CLDMASK ,NDVI ,BAND1 ,BAND2 ,BAND3 ,BAND4,AOD):
print ("processing:"+ a)
arcpy.AddMessage("processing:{}".format(b.split('\\')[4][9:30]))
name = c.split("\\")
filename=name[6][9:33]
print filename
arcpy.AddMessage("processing:{}".format(d.split('\\')[6][9:33]))
arcpy.AddMessage("processing:{}".format(e.split('\\')[6][9:30]))
arcpy.AddMessage("processing:{}".format(f.split('\\')[6][9:33]))
print ("processing:"+ g)
#######################################################################
setnull =arcpy.gp.SetNull_sa(g,g, "in_memory/dat", "\"Value\" = -9999")
ras=arcpy.Raster(setnull)
Data=(ras-add_offset)*Scale_factor
da = Con((Data >= 0.0) & (Data <= 0.1),1)
# Process: Extract by Cloud_Mask
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = "MAXOF"
ndvi_msk=arcpy.gp.ExtractByMask_sa(b, a)
Band1_mask=arcpy.gp.ExtractByMask_sa(c, a)
Band2_mask=arcpy.gp.ExtractByMask_sa(d, a)
Band3_mask=arcpy.gp.ExtractByMask_sa(e, a)
Band4_mask=arcpy.gp.ExtractByMask_sa(f, a)
arcpy.env.cellSize = tempEnvironment0
# Process: Extract by Mask using AOD less than 0.1 value
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = cellsize
ndvi_msk1=arcpy.gp.ExtractByMask_sa(ndvi_msk, da)
Band1_mask1=arcpy.gp.ExtractByMask_sa(Band1_mask, da)
Band2_mask2=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
Band3_mask3=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
Band4_mask4=arcpy.gp.ExtractByMask_sa(Band2_mask, da)
arcpy.env.cellSize = tempEnvironment0
try:
# Raster to point
field ="VALUE"
Point=arcpy.RasterToPoint_conversion(ndvi_msk1,"in_memory/fileroot",field)
shape=arcpy.sa.ExtractMultiValuesToPoints(Point,[[ndvi_msk1,"NDVI"],[Band1_mask1,"Band1"],[Band2_mask2,"Band2"],
[Band3_mask3 ,"Band3"],[Band4_mask4,"Band4"]], "NONE")
#Add Two Fields Name and Name2
[arcpy.AddField_management(shape,field_name, "TEXT", field_length = 50)
for field_name in ["Name", "Name2"]]
arcpy.AddMessage("Successfully Added NAME fields ")
fieldss = ['Band1','Name']
with arcpy.da.UpdateCursor(shape, fieldss) as cursor:
for row in cursor:
if (row[0] > 0.02 and row[0] < 0.06):
row[1] = 'AAA'
print ("processing1:""{0:.8f}, {1}".format(row[0], row[1]))
elif (row[0] >= 0.06 and row[0] < 0.09):
row[1] = 'BBB'
# print ("processing1:""{0:.8f}, {1}".format(row[0], row[1]))
elif (row[0] >= 0.09):
row[1] = 'CCC'
else:
row[1] = 'NA'
# Update the cursor with the updated list
cursor.updateRow(row)
# Process: Select Layer By Attribute for separate the catagories "AAA","BBB,"CCC"
arcpy.MakeFeatureLayer_management(shape,"lyr1")
AAA = arcpy.SelectLayerByAttribute_management("lyr1", "NEW_SELECTION", "\"Name\" = 'AAA'")
print "Successfully Process : Select Layer By Attribute for AAA "
arcpy.MakeFeatureLayer_management(shape,"lyr2")
BBB = arcpy.SelectLayerByAttribute_management("lyr2", "NEW_SELECTION", "\"Name\" = 'BBB'")
print "Successfully Process : Select Layer By Attribute for BBB "
arcpy.MakeFeatureLayer_management(shape,"lyr3")
CCC = arcpy.SelectLayerByAttribute_management("lyr3", "NEW_SELECTION", "\"Name\" = 'CCC'")
print "Successfully Process : Select Layer By Attribute for CCC "
# Writing tables for categories("AAA","BBB,"CCC") and individual category have three Land type
# that will be save in different folder and their individual name
#------------------First we write category "AAA" and their indiviual land type--------------------------------------#
def geoprocess1(base_path,category,file_name,file):
try:
land_use='LAND_10'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
try:
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
except OSError:
print ("Creation of the directory failed" )
else:
print ("Successfully created the directory" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['grid_code', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 10")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(file, fields) as cursor:
for row in cursor:
if (row[0] > 0.10 and row[0] < 0.20):
row[5] = 'landuse_10'
print ("processing_A:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 10 " )
print('Sucess: geoprocess1 complete!')
except Exception as err:
print('Error: unable to complete geoprocess1')
print(err)
def geoprocess2(base_path,category,file_name,file):
try:
land_use='LAND_20'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
try:
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
except OSError:
print ("Creation of the directory failed" )
else:
print ("Successfully created the directory" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['grid_code', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 20")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(file, fields) as cursor:
for row in cursor:
if (row[0] >= 0.20 and row[0] < 0.50):
row[5] = 'landuse_20'
print ("processing_B:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 20 " )
print('Sucess: geoprocess2 complete!')
except Exception as err:
print('Error: unable to complete geoprocess2')
print(err)
def geoprocess3(base_path,category,file_name,file):
try:
land_use='LAND_30'
outdir = os.path.join(base_path,category,land_use)
if not os.path.exists(outdir):
try:
print('\nCreating new output directory!\n'+outdir+'\n')
os.makedirs(outdir)
except OSError:
print ("Creation of the directory failed" )
else:
print ("Successfully created the directory" )
ras_name = os.path.join(outdir,("{0}_{1}{2}_{3}".format(category,file_name,land_use,".txt")))
fields = ['grid_code', 'Band1', 'Band2','Band3','Name',"Name2"]
with open(ras_name, 'wb') as txtfile:
arcpy.AddMessage("fields Writing For landuse 30")
txtfile.write(" {0} {1} {2} {3} {4} {5}\n".format(fields[0],fields[1],fields[2],fields[3],fields[4],fields[5]))
with arcpy.da.UpdateCursor(file, fields) as cursor:
for row in cursor:
if (row[0] >= 0.50):
row[5] = 'landuse_30'
print ("processing_C:""{0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.write(" {0:.8f} {1:.8f} {2:.8f} {3:.8f} {4} {5}\n".format(row[0],row[1],row[2], row[3],row[4],row[5]))
txtfile.close()
print ("Successfully save the landuse 30 " )
print('Sucess: geoprocess3 complete!')
except Exception as err:
print('Error: unable to complete geoprocess3')
print(err)
def main(base_path,category,file_name,file):
geoprocess1(base_path,category,file_name,file)
geoprocess2(base_path,category,file_name,file)
geoprocess3(base_path,category,file_name,file)
#category="AAA"
file_name=filename
base_path = r"F:\\DB_test_data\\VAR4\\"
if __name__ == "__main__":
#category="AAA"
file_name="MOD022"
base_path = r"F:\\DB_test_data\\VAR4\\"
try:
t1=time.time()
main(base_path,"AAA",file_name,AAA)
main(base_path,"BBB",file_name,BBB)
main(base_path,"CCC",file_name,CCC)
print "serial processing took:",time.time()-t1
print "serial processing is OK"
except Exception as e:
print "Error in serial processing"
print(err)
try:
t2=time.time()
pool = multiprocessing.Pool(processes=4)
A=pool.map(main,(base_path,"AAA",file_name,AAA))
B=pool.map(main,(base_path,"BBB",file_name,BBB))
C=pool.map(main,(base_path,"CCC",file_name,CCC))
A.close()
B.close()
C.close()
A.join()
B.join()
C.join()
print "Pool took",time.time()-t2
print "multiprocessing is OK"
except Exception as e:
print "Error in multiprocessing "
#print(err)
print arcpy.GetMessages()
print 'finished run: %s\n\n' % (datetime.datetime.now() - start)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Again... does it work for 1 set and what is the output? Does the above error out? Does it produce anything or does it fail?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
First of all, the code does not show any error But when I include the multiprocessing function is not working. In every category ("AAA","BBB","CCC") have Ten landuse_type initially I write three types (geoprocess1 to 3) it needs to write more seven processes then the code will be bigger that's why I need to reduce this code. I previously uploaded my folder structure of one category( "AAA") and their output text file. Dan Patterson