- Home
- :
- All Communities
- :
- Developers
- :
- Python
- :
- Python Questions
- :
- RuntimeError: ERROR 999998: Unexpected Error in Se...
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
RuntimeError: ERROR 999998: Unexpected Error in Set null function
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
I am trying to write one code where I use set null function for different images and also use a simple math function for scaling the images. But when I run this code after three processes it shows
Traceback (most recent call last):
File "F:\DB_test_data\python_script\FINAL_DB.py", line 192, in <module>
arcpy.CopyRaster_management(toa_b2,OutRaster)
File "C:\Program Files (x86)\ArcGIS\Desktop10.6\ArcPy\arcpy\management.py", line 14587, in CopyRaster
raise e
RuntimeError: ERROR 999998: Unexpected Error.here is my code
outdir="F:\\DB_test_data\\TEST_RAY\\TEST1\\c\\"
for a,b,c,d,e,f,g,h,i,j,k,l in zip (CLDMASK,AOD,TOA_B1,TOA_B2,Height,SenZen,SenAzm,SolZen,SolAzm,Sur_B1,Sur_B2,Sur_B3):
print ("processing_a:"+ a)
print ("processing_b:"+ b)
print ("processing_c:"+ c)
print ("processing_d:"+ d)
print ("processing_e:"+ e)
print ("processing_f:"+ f)
print ("processing_g:"+ g)
print ("processing_h:"+ h)
print ("processing_i:"+ i)
print ("processing_j:"+ j)
print ("processing_k:"+ k)
print ("processing_l:"+ l)
#######################################################################
#Cloud Mask scalling
Scale_factor1 = float(1.0)
add_offset1 = float(0.0)
setnull1 =arcpy.gp.SetNull_sa(a,a, "in_memory/dat1", "\"Value\" <= 0")
ras1=arcpy.Raster(setnull1)
cldmask=(ras1-add_offset1)*Scale_factor1
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("cldmask"))
#print OutRaster
arcpy.CopyRaster_management(cldmask,OutRaster)
arcpy.Delete_management("in_memory/dat1")
#AOD scalling and set lessthan 0.1 AOD
Scale_factor2 = float(0.0010000000474974513)
add_offset2 = float(0.0)
setnull2 =arcpy.gp.SetNull_sa(b,b, "in_memory/dat2", "\"Value\" = -9999")
ras2=arcpy.Raster(setnull2)
aod=(ras2-add_offset2)*Scale_factor2
aod_1 = Con((aod >= 0.0) & (aod <= 0.1),1)
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("aod_1"))
#print OutRaster
arcpy.CopyRaster_management(aod_1,OutRaster)
arcpy.Delete_management("in_memory/dat2")
#TOA_B1 scalling
Scale_factor3 = float(0.000053811826)
add_offset3 = float(-0.0)
setnull3 =arcpy.gp.SetNull_sa(c,c, "in_memory/dat3", "\"Value\" = 65535")
ras3=arcpy.Raster(setnull3)
toa_b1=(ras3-(add_offset3))*Scale_factor3
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("toa_b1"))
#print OutRaster
arcpy.CopyRaster_management(toa_b1,OutRaster)
arcpy.Delete_management("in_memory/dat3")
#TOA_B2 Scalling
Scale_factor4 = float(0.00003255546)
add_offset4 = float(-0.0)
setnull4 =arcpy.gp.SetNull_sa(d,d, "in_memory/dat4", "\"Value\" = 65535")
ras4=arcpy.Raster(setnull4)
toa_b2=(ras4-(add_offset4))*Scale_factor4
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("toa_b2"))
#print OutRaster
arcpy.CopyRaster_management(toa_b2,OutRaster)
arcpy.Delete_management("in_memory/dat4")
#Height
setnull =arcpy.gp.SetNull_sa(e,e, "in_memory/dat5", "\"Value\" = -32767")
ras=arcpy.Raster(setnull)
height=ras
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("height"))
#print OutRaster
arcpy.CopyRaster_management(height,OutRaster)
arcpy.Delete_management("in_memory/dat5")
#Sensor Zenith angle
Scale_factor6 = float(0.01)
add_offset6 = float(0.0)
setnull6 =arcpy.gp.SetNull_sa(f,f, "in_memory/dat6", "\"Value\" = -32767")
ras6=arcpy.Raster(setnull6)
senzen=(ras6-add_offset6)*Scale_factor6
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("senzen"))
#print OutRaster
arcpy.CopyRaster_management(senzen,OutRaster)
arcpy.Delete_management("in_memory/dat6")
#Sensor azimuth angle
setnull7 =arcpy.gp.SetNull_sa(g,g, "in_memory/dat7", "\"Value\" = -32767")
ras7=arcpy.Raster(setnull7)
senazm=(ras7-add_offset6)*Scale_factor6
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("senazm"))
#print OutRaster
arcpy.CopyRaster_management(senazm,OutRaster)
arcpy.Delete_management("in_memory/dat7")
#solar zenithal angle
setnull8 =arcpy.gp.SetNull_sa(h,h, "in_memory/dat8", "\"Value\" = -32767")
ras8=arcpy.Raster(setnull8)
solzen=(ras8-add_offset6)*Scale_factor6
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("solzen"))
#print OutRaster
arcpy.CopyRaster_management(solzen,OutRaster)
arcpy.Delete_management("in_memory/dat8")
#solar azimuth angle
setnull9 =arcpy.gp.SetNull_sa(i,i, "in_memory/dat9", "\"Value\" = -32767")
ras9=arcpy.Raster(setnull9)
solazm=(ras9-add_offset6)*Scale_factor6
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("solazm"))
#print OutRaster
arcpy.CopyRaster_management(solazm,OutRaster)
arcpy.Delete_management("in_memory/dat9")
#(MODO9)surface reflectance band 1
Scale_factor_10 = float(0.0001)
add_offset_10 = float(0.0)
setnull_10 =arcpy.gp.SetNull_sa(j,j, "in_memory/dat10", "\"Value\" = -28672")
ras_10=arcpy.Raster(setnull_10)
sur_b1=(ras_10-add_offset_10)*Scale_factor_10
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("sur_b1"))
#print OutRaster
arcpy.CopyRaster_management(sur_b1,OutRaster)
arcpy.Delete_management("in_memory/dat10")
#(MODO9)surface reflectance band 2
setnull_11 =arcpy.gp.SetNull_sa(k,k, "in_memory/dat11", "\"Value\" = -28672")
ras_11=arcpy.Raster(setnull_11)
sur_b2=(ras_11-add_offset_10)*Scale_factor_10
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("sur_b2"))
#print OutRaster
arcpy.CopyRaster_management(sur_b2,OutRaster)
arcpy.Delete_management("in_memory/dat11")
#(MODO9)surface reflectance band 3
setnull_12 =arcpy.gp.SetNull_sa(l,l, "in_memory/dat12", "\"Value\" = -28672")
ras_12=arcpy.Raster(setnull_12)
sur_b3=(ras_12- add_offset_10)*Scale_factor_10
OutRaster = os.path.join(outdir,'MOd0.{0}.img'.format("sur_b3"))
#print OutRaster
arcpy.CopyRaster_management(sur_b3,OutRaster)
arcpy.Delete_management("in_memory/dat12")
#############################################################################
# # Process: Extract by Cloud_Mask
# tempEnvironment0 = arcpy.env.cellSize
# arcpy.env.cellSize = "MAXOF"
# toa_bb1=arcpy.gp.ExtractByMask_sa(toa_b1, cldmask)
# toa_b22=arcpy.gp.ExtractByMask_sa(toa_b2, cldmask)
# heightt=arcpy.gp.ExtractByMask_sa(height, cldmask)
# senzenn=arcpy.gp.ExtractByMask_sa(senzen, cldmask)
# senazmm=arcpy.gp.ExtractByMask_sa(senazm, cldmask)
# solzenn=arcpy.gp.ExtractByMask_sa(solzen, cldmask)
# solazmm=arcpy.gp.ExtractByMask_sa(solazm, cldmask)
# sur_b11=arcpy.gp.ExtractByMask_sa(sur_b1, cldmask)
# sur_b22=arcpy.gp.ExtractByMask_sa(sur_b2, cldmask)
# sur_b33=arcpy.gp.ExtractByMask_sa(sur_b3, cldmask)
# arcpy.env.cellSize = tempEnvironment0
# # Process: Extract by Mask using AOD less than 0.1 value
# tempEnvironment0 = arcpy.env.cellSize
# arcpy.env.cellSize = cellsize
# toa_b1=arcpy.gp.ExtractByMask_sa(toa_bb1, aod_1)
# toa_b2=arcpy.gp.ExtractByMask_sa(toa_bb2, aod_1)
# height=arcpy.gp.ExtractByMask_sa(heightt, aod_1)
# senzen=arcpy.gp.ExtractByMask_sa(senzenn, aod_1)
# senazm=arcpy.gp.ExtractByMask_sa(senazmm, aod_1)
# solzen=arcpy.gp.ExtractByMask_sa(solzenn, aod_1)
# solazm=arcpy.gp.ExtractByMask_sa(solazmm, aod_1)
# sur_b1=arcpy.gp.ExtractByMask_sa(sur_b11, aod_1)
# sur_b2=arcpy.gp.ExtractByMask_sa(sur_b22, aod_1)
# sur_b3=arcpy.gp.ExtractByMask_sa(sur_b33, aod_1)
# arcpy.env.cellSize = tempEnvironment0
#############################################################################
#Calculate the scattering angle from Resample MOD03 products
#Css = Cos(S2 * !pi / 180.D) IDL
#Css = np.cos(S2 * np.pi / 180) Python
# CosSenZ = np.Cos(senzen * np.pi / 180)
# CosSolZ = np.Cos(solzen * np.pi / 180)
# SinSenZ = np.Sin(senzen * np.pi / 180)
# SinSolZ = np.Sin(solzen * np.pi / 180)
# RelAzm = np.abs((senazm ) - (solazm))
# index = np.where(RelAzm > 180.0)
# RelAzm[index] = 360.0- RelAzm[index]
# index = np.where(RelAzm <= 180.0)
# RelAzm[index] = 180.0- RelAzm[index]
# CosRe = np.Cos(np.radians(RelAzm))
# SctAgl = np.acos((-CosSolZ * CosSenZ) + ((SinSolZ * SinSenZ) * CosRelA))
# #Ray_correction for TOA_BAND_1 and TOA_BAND_2
# #TOA_BAND_1
# B1_ROD = (0.00864 + (0.0000065)) * (0.67)**(-(3.916 + (0.074 * 0.67)+ (0.05/0.67)))#for RED band, please change 0.67 for other bands wavelength
# Pr1 = (3./16.*np.pi) * (1 + (np.cos(SctAgl) * np.cos(SctAgl)))
# wr1 = 1.0
# RayRef_B1 = (wr1 * B1_ROD * Pr1 ) / (4.0 * (CosSolZ * CosSenZ))
# RCR_B1 = toa_b1 - RayRef_B1
# #TOA_BAND_2
# B2_ROD = (0.00864 + (0.0000065)) * (0.86)**(-(3.916 + (0.074 * 0.86)+ (0.05/0.86)))#for Near Infrared (NIR) band, please change 0.86 for other bands wavelength
# Pr2 = (3./16.*np.pi) * (1 + (np.cos(SctAgl) * np.cos(SctAgl)))
# wr2 = 1.0
# RayRef_B2 = (wr2 * B2_ROD * Pr2 ) / (4.0 * (CosSolZ * CosSenZ))
# RCR_B2 = toa_b2 - RayRef_B2
#Calculating NDVI float raster
# ndvi_raster = Divide(Float(Raster(toa_b2) - Raster(toa_b1)), Float(Raster(toa_b2) + Raster(toa_b1)))
# ndvi = (Float(toa_b2) - toa_b1) / (Float(toa_b2) + toa_b1)
print arcpy.GetMessages()
print 'finished run: %s\n\n' % (datetime.datetime.now() - start)
I can't understand why it shows this error.
please help me, anyone, to resolve this matter.
And have any way to use a "foor loop" for set null function of different images?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi BIJOY GAYEN ,
I haven't been able to find the time yet to validate the script with your data. I did download the data, but I still need to validate what might be causing the error.
Hopefully, I will be able to do this soon, but I have a long list of things that I have to do, so I can't promise you that I will be able to do this today or tomorrow.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi BIJOY GAYEN ,
In order for you to continue, a view pointers:
- On line 316, you are using np.acos, this should be np.arccos
- On line 316 you also use a numpy array variable called "cosRelA". This one does not exist, perhaps it is "cosRe" created on row 314
- On line 324 you are creating "toa_b2_na" when this should be "toa_b1_na" created based on "toa_b1" (I guess)
- On line 325 your script will produce an error since the numpy arrays have different dimensions: "ValueError: operands could not be broadcast together with shapes (202,1007) (203,1008)". Make sure that all raster have the same extent and pixel size before converting them to numpy arrays and combining them (use a proper snap raster, extent and pixelsize in your settings)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hello Xander Bakker sir,
I rechecked my code as per your suggestion, But when I try to save the output SctAgl and toa_b1 after some processes this code gave me some error in output (the output gave me blank result with unwanted value range in two images)
SctAgl:
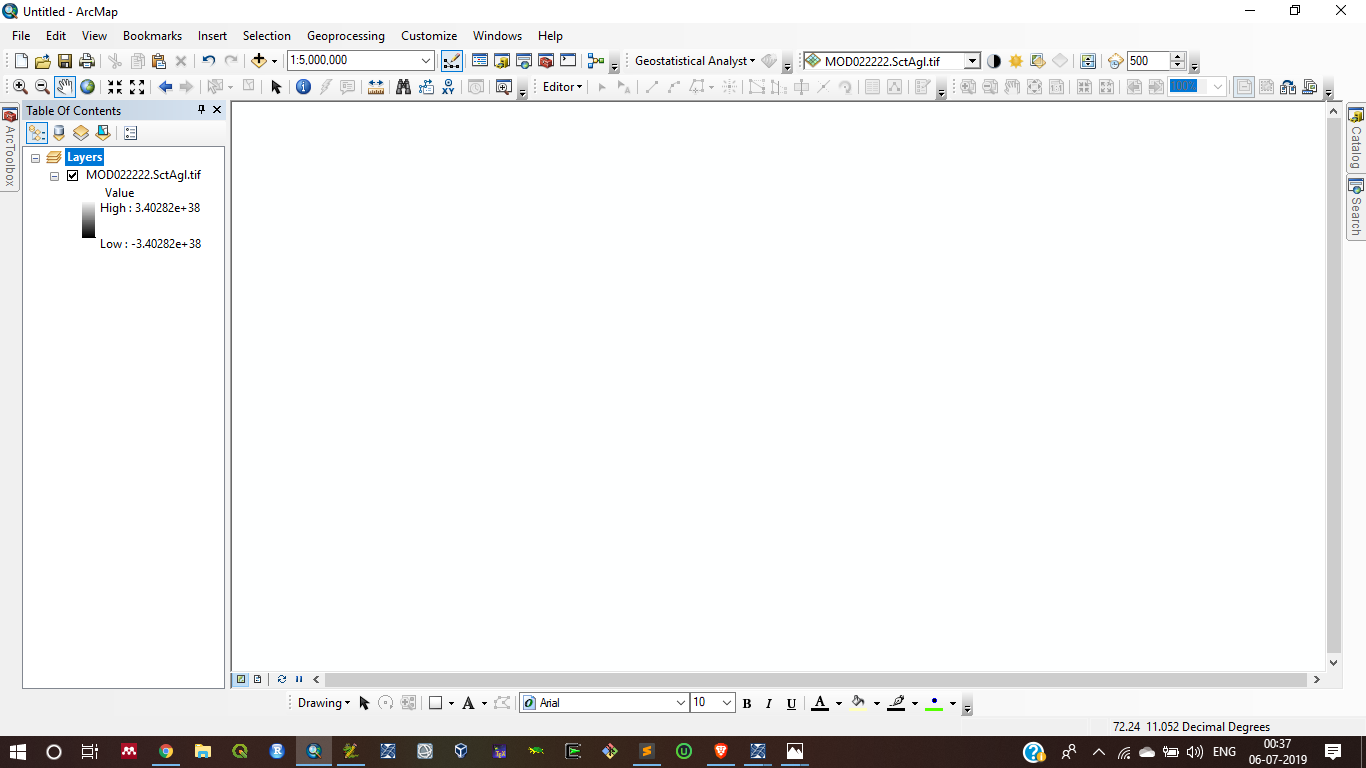
Toa_B1:
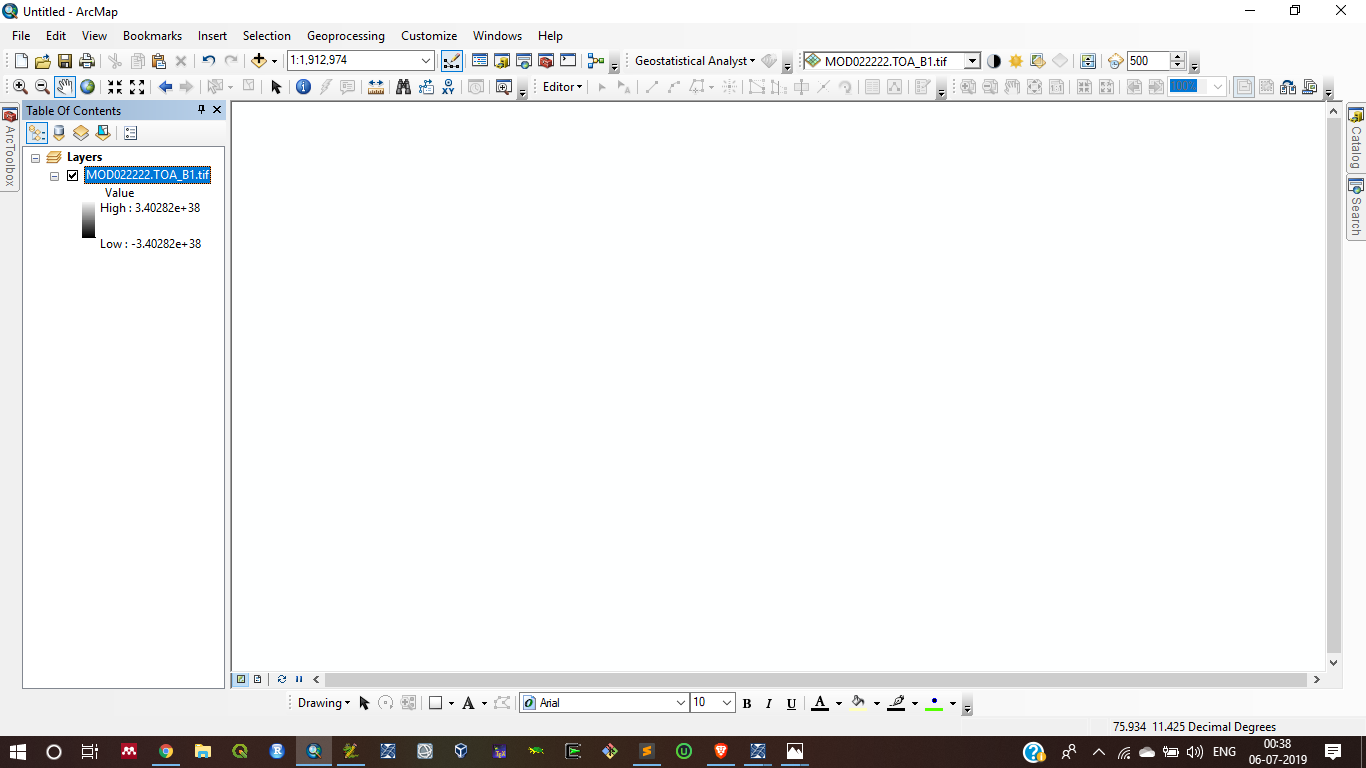
Second, when I run this code it shows some unwanted messages, I cannot understand why these messages occur
F:\DB_test_data\python_script\FINAL_DB.py:298: RuntimeWarning: overflow encountered in multiply
cosSenZ = np.cos(senzen_na * np.pi / 180)
F:\DB_test_data\python_script\FINAL_DB.py:298: RuntimeWarning: invalid value encountered in cos
cosSenZ = np.cos(senzen_na * np.pi / 180)
F:\DB_test_data\python_script\FINAL_DB.py:300: RuntimeWarning: overflow encountered in multiply
cosSolZ = np.cos(cosSolZ_na * np.pi / 180)
F:\DB_test_data\python_script\FINAL_DB.py:300: RuntimeWarning: invalid value encountered in cos
cosSolZ = np.cos(cosSolZ_na * np.pi / 180)
F:\DB_test_data\python_script\FINAL_DB.py:302: RuntimeWarning: overflow encountered in multiply
sinSenZ = np.sin(sinSenZ_na * np.pi / 180)
F:\DB_test_data\python_script\FINAL_DB.py:302: RuntimeWarning: invalid value encountered in sin
sinSenZ = np.sin(sinSenZ_na * np.pi / 180)
F:\DB_test_data\python_script\FINAL_DB.py:304: RuntimeWarning: overflow encountered in multiply
sinSolZ = np.sin(sinSolZ_na * np.pi / 180)
F:\DB_test_data\python_script\FINAL_DB.py:304: RuntimeWarning: invalid value encountered in sin
sinSolZ = np.sin(sinSolZ_na * np.pi / 180)Third things, when I try to run this code with row data (whole India) after some processing it shows memory issue
sinSenZ_na = arcpy.RasterToNumPyArray(senzen)
File "C:\Program Files (x86)\ArcGIS\Desktop10.6\ArcPy\arcpy\__init__.py", line 2275, in RasterToNumPyArray
return _RasterToNumPyArray(*args, **kwargs)
MemoryErrorThere is no option, without conversion (Raster To Numpy and then Numpy To Raster) to write the code for raster data analysis directly use cos, sin, subtract, plus, minus, division function, etc. I am totally confused to write this code. Give any solution regarding this issue.
Dan Patterson Joe BorgioneJoshua BixbyLuke Webb
My_full_code:
#-------------------------------------------------------------------------------
# Name: module1
# Purpose:
#
# Author: HONEY
#
# Created: 18-06-2019
# Copyright: (c) HONEY 2019
# Licence: <your licence>
#-------------------------------------------------------------------------------
import datetime
start = datetime.datetime.now()
print 'start run: %s\n' % (start)
import arcpy ,os ,sys,csv,errno
from arcpy import env
from arcpy.sa import *
import datetime
import re
import glob
import itertools
import math
import numpy as np
arcpy.CheckOutExtension('Spatial')
arcpy.env.overwriteOutput = True
cellsize = "F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip\\MOD02HKM_A2017001_0530_NDVI_AA.img"
#Call cloud mask images from directory
d1="F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip"
CLDMASK = glob.glob(d1 + os.sep + "*.Aerosol_Cldmask_Land_Ocean-Aerosol_Cldmask_Land_Ocean.tif")
CLDMASK.sort()
if len(CLDMASK) == 0:
print 'Could not open the CLDMASK raster files'
sys.exit(1)
else:
print 'The CLDMASK raster files was opened successfully'
#Call AOD images from directory
d2 = r"F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip"
AOD = glob.glob(d2 + os.sep + "*Corrected_Optical_Depth_Land_2-Corrected_Optical_Depth_Land.tif")
AOD.sort()
if len(AOD) == 0:
print 'Could not open the AOD raster files'
sys.exit(1)
else:
print 'The AOD raster files was opened successfully'
#Call MOD02HKM_TOA-B1_TOA-B2 images from directory
d3="F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip"
TOA_B1 = glob.glob(d3 + os.sep + "*EV_500_RefSB_1-EV_500_RefSB.tif")
TOA_B1.sort()
if len(TOA_B1) == 0:
print 'Could not open the TOA_B1 raster files'
sys.exit(1)
else:
print 'The TOA_B1 raster files was opened successfully'
TOA_B2 = glob.glob(d3 + os.sep + "*EV_500_RefSB_2-EV_500_RefSB.tif")
TOA_B2.sort()
if len(TOA_B2) == 0:
print 'Could not open the TOA_B2 raster files'
sys.exit(1)
else:
print 'The TOA_B2 raster files was opened successfully'
#Call MOD03 (Height,SenZen,SenAzm,SolZen,SolAzm) images from directory
d4="F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip"
Height = glob.glob(d4 + os.sep + "*.Height-Height.tif")
Height.sort()
if len(Height) == 0:
print 'Could not open the Height raster files'
sys.exit(1)
else:
print 'The Height raster files was opened successfully'
SenZen = glob.glob(d4 + os.sep + "*.SensorZenith-SensorZenith.tif")
SenZen.sort()
if len(SenZen) == 0:
print 'Could not open the SenZen raster files'
sys.exit(1)
else:
print 'The SenZen raster files was opened successfully'
SenAzm = glob.glob(d4 + os.sep + "*.SensorAzimuth-SensorAzimuth.tif")
SenAzm.sort()
if len(SenAzm) == 0:
print 'Could not open the SenAzm raster files'
sys.exit(1)
else:
print 'The SenAzm raster files was opened successfully'
SolZen = glob.glob(d4 + os.sep + "*.SolarZenith-SolarZenith.tif")
SolZen.sort()
if len(SolZen) == 0:
print 'Could not open the SolZen raster files'
sys.exit(1)
else:
print 'The SolZen raster files was opened successfully'
SolAzm = glob.glob(d4 + os.sep + "*.SolarAzimuth-SolarAzimuth.tif")
SolAzm.sort()
if len(SolAzm) == 0:
print 'Could not open the SolAzm raster files'
sys.exit(1)
else:
print 'The SolAzm raster files was opened successfully'
#Call MOD09 images from directory
d5="F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip"
Sur_B1 = glob.glob(d5 + os.sep + "*.500m_Surface_Reflectance_Band_1-500m_Surface_Reflectance_Band_1.tif")
Sur_B1.sort()
if len(Sur_B1) == 0:
print 'Could not open the Sur_B1 raster files'
sys.exit(1)
else:
print 'The Sur_B1 raster files was opened successfully'
Sur_B2 = glob.glob(d5 + os.sep + "*.500m_Surface_Reflectance_Band_2-500m_Surface_Reflectance_Band_2.tif")
Sur_B2.sort()
if len(Sur_B2) == 0:
print 'Could not open the Sur_B2 raster files'
sys.exit(1)
else:
print 'The Sur_B2 raster files was opened successfully'
Sur_B3 = glob.glob(d5 + os.sep + "*.500m_Surface_Reflectance_Band_3-500m_Surface_Reflectance_Band_3.tif")
Sur_B3.sort()
if len(Sur_B3) == 0:
print 'Could not open the Sur_B3 raster files'
sys.exit(1)
else:
print 'The Sur_B3 raster files was opened successfully'
Land_cover="F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip\\LAND_COVER_TYPE_1_Grid_2D.img"
if len(Land_cover) == 0:
print 'Could not open the Land_cover raster files'
sys.exit(1)
else:
print 'The Land_cover raster files was opened successfully'
outdir="F:\\DB_test_data\\TEST_RAY\\TEST1\\c\\d\\"
cellsize = "F:\\DB_test_data\\VAR2\\TEST_DATA\\Clip\\MOD02HKM_A2017001_0530_NDVI_AA.img"
for i in range(len(CLDMASK)):
AA=CLDMASK[i].split("\\")[5][0:114]
print ("processing_a:"+ AA)
BB=AOD[i].split("\\")[9][0:120]
print ("processing_b:"+ BB)
CC=TOA_B1[i].split("\\")[5][0:88]
print ("processing_c:"+ CC)
DD=TOA_B2[i].split("\\")[5][0:88]
print ("processing_d:"+ DD)
EE=Height[i].split("\\")[5][0:71]
print ("processing_e:"+ EE)
FF=SenZen[i].split("\\")[5][0:83]
print ("processing_f:"+ FF)
GG=SenAzm[i].split("\\")[5][0:85]
print ("processing_g:"+ GG)
HH=SolZen[i].split("\\")[5][0:81]
print ("processing_h:"+ HH)
II=SolAzm[i].split("\\")[5][0:83]
print ("processing_i:"+ II)
JJ=Sur_B1[i].split("\\")[5][0:121]
print ("processing_j:"+ JJ)
KK=Sur_B2[i].split("\\")[5][0:121]
print ("processing_k:"+ KK)
LL=Sur_B3[i].split("\\")[5][0:121]
print ("processing_l:"+ LL)
#######################################################################
#Cloud Mask scalling
Scale_factor1 = float(1.0)
add_offset1 = float(0.0)
whereClause = "VALUE <= 0"
# Execute SetNull
outSetNull = SetNull(CLDMASK[i], CLDMASK[i], whereClause)
cldmask=(outSetNull-add_offset1)*Scale_factor1
#AOD scalling
Scale_factor2 = float(0.0010000000474974513)
add_offset2 = float(0.0)
whereClause= "VALUE = -9999"
# Execute SetNull
outSetNull2 = SetNull(AOD[i],AOD[i], whereClause)
aod=(outSetNull2-add_offset2)*Scale_factor2
aod_1 = Con((aod >= 0.0) & (aod <= 0.1),1)
#TOA_B1 Scalling
Scale_factor3 = float(0.000053811826)
add_offset3 = float(-0.0)
whereClause= "VALUE = 65535"
# Execute SetNull
outSetNull3 = SetNull(TOA_B1[i],TOA_B1[i], whereClause)
toa_b1=(outSetNull3-add_offset3)*Scale_factor3
#TOA_B2 Scalling
Scale_factor4 = float(0.00003255546)
add_offset4 = float(-0.0)
whereClause= "VALUE = 65535"
# Execute SetNull
outSetNull4 = SetNull(TOA_B2[i],TOA_B2[i], whereClause)
toa_b2=(outSetNull4-add_offset4)*Scale_factor4
#Height
Scale_factor5 = float(1.0)
add_offset5 = float(-0.0)
whereClause = "VALUE = -32767"
# Execute SetNull
outSetNull5 = SetNull(Height[i],Height[i], whereClause)
height=(outSetNull5-add_offset5)*Scale_factor5
#Sensor Zenith angle
Scale_factor6 = float(0.01)
add_offset6 = float(0.0)
whereClause = "VALUE = -32767"
# Execute SetNull
outSetNull6 = SetNull(SenZen[i],SenZen[i], whereClause)
senzen=(outSetNull6-add_offset6)*Scale_factor6
#Sensor azimuth angle
Scale_factor7 = float(0.01)
add_offset7 = float(0.0)
whereClause = "VALUE = -32767"
# Execute SetNull
outSetNull7 = SetNull(SenAzm[i],SenAzm[i], whereClause)
senazm=(outSetNull7-add_offset7)*Scale_factor7
#solar Zenith angle
Scale_factor8 = float(0.01)
add_offset8 = float(0.0)
whereClause = "VALUE = -32767"
# Execute SetNull
outSetNull8 = SetNull(SolZen[i],SolZen[i], whereClause)
solzen=(outSetNull8-add_offset8)*Scale_factor8
#solar azimuth angle
Scale_factor9 = float(0.01)
add_offset9 = float(0.0)
whereClause = "VALUE = -32767"
# Execute SetNull
outSetNull9 = SetNull(SolAzm[i],SolAzm[i], whereClause)
solazm=(outSetNull9-add_offset9)*Scale_factor9
# OutRaster = os.path.join(outdir,'MOD04_L22.{0}.tif'.format("solazm"))
# solazm.save(OutRaster)
#(MODO9)surface reflectance band 1
Scale_factor_10 = float(0.0001)
add_offset_10 = float(0.0)
whereClause = "VALUE = -28672"
# Execute SetNull
outSetNull10 = SetNull(Sur_B1[i],Sur_B1[i], whereClause)
sur_b1=(outSetNull10-add_offset_10)*Scale_factor_10
#(MODO9)surface reflectance band 2
Scale_factor_11 = float(0.0001)
add_offset_11 = float(0.0)
whereClause = "VALUE = -28672"
# Execute SetNull
outSetNull11 = SetNull(Sur_B2[i],Sur_B2[i], whereClause)
sur_b2=(outSetNull11-add_offset_11)*Scale_factor_11
#(MODO9)surface reflectance band 3
Scale_factor_12 = float(0.0001)
add_offset_12 = float(0.0)
whereClause = "VALUE = -28672"
# Execute SetNull
outSetNull12 = SetNull(Sur_B3[i],Sur_B3[i], whereClause)
sur_b3=(outSetNull12-add_offset_12)*Scale_factor_12
# Process: Extract by Cloud_Mask
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = "MAXOF"
toa_bb1=arcpy.sa.ExtractByMask(toa_b1, cldmask)
toa_bb2=arcpy.sa.ExtractByMask(toa_b2, cldmask)
heightt=arcpy.sa.ExtractByMask(height, cldmask)
senzenn=arcpy.sa.ExtractByMask(senzen, cldmask)
senazmm=arcpy.sa.ExtractByMask(senazm, cldmask)
solzenn=arcpy.sa.ExtractByMask(solzen, cldmask)
solazmm=arcpy.sa.ExtractByMask(solazm, cldmask)
sur_b11=arcpy.sa.ExtractByMask(sur_b1, cldmask)
sur_b22=arcpy.sa.ExtractByMask(sur_b2, cldmask)
sur_b33=arcpy.sa.ExtractByMask(sur_b3, cldmask)
arcpy.env.cellSize = tempEnvironment0
# # Process: Extract by Mask using AOD less than 0.1 value
tempEnvironment0 = arcpy.env.cellSize
arcpy.env.cellSize = cellsize
toa_b1=arcpy.sa.ExtractByMask(toa_bb1, aod_1)
toa_b2=arcpy.sa.ExtractByMask(toa_bb2, aod_1)
height=arcpy.sa.ExtractByMask(heightt, aod_1)
senzen=arcpy.sa.ExtractByMask(senzenn, aod_1)
senazm=arcpy.sa.ExtractByMask(senazmm, aod_1)
solzen=arcpy.sa.ExtractByMask(solzenn, aod_1)
solazm=arcpy.sa.ExtractByMask(solazmm, aod_1)
sur_b1=arcpy.sa.ExtractByMask(sur_b11, aod_1)
sur_b2=arcpy.sa.ExtractByMask(sur_b22, aod_1)
sur_b3=arcpy.sa.ExtractByMask(sur_b33, aod_1)
arcpy.env.cellSize = tempEnvironment0
#############################################################################
#Calculate the scattering angle from Resample MOD03 products
senzen_na = arcpy.RasterToNumPyArray(senzen)
cosSenZ = np.cos(senzen_na * np.pi / 180)
cosSolZ_na = arcpy.RasterToNumPyArray(solzen)
cosSolZ = np.cos(cosSolZ_na * np.pi / 180)
sinSenZ_na = arcpy.RasterToNumPyArray(senzen)
sinSenZ = np.sin(sinSenZ_na * np.pi / 180)
sinSolZ_na = arcpy.RasterToNumPyArray(solzen)
sinSolZ = np.sin(sinSolZ_na * np.pi / 180)
senazm_na = arcpy.RasterToNumPyArray(senazm)
solazm_na = arcpy.RasterToNumPyArray(solazm)
RelAzm = np.abs((senazm_na ) - (solazm_na))
index = np.where(RelAzm > 180.0)
RelAzm[index] = 360.0- RelAzm[index]
index = np.where(RelAzm <= 180.0)
RelAzm[index] = 180.0- RelAzm[index]
cosRelA = np.cos(np.radians(RelAzm))
SctAgl = np.arccos((-cosSolZ * cosSenZ) + ((sinSolZ * sinSenZ) * cosRelA))
dscc=arcpy.Describe(senazm)
srr=dscc.SpatialReference
extt=dscc.Extent
lll=arcpy.Point(extt.XMin,extt.YMin)
noDataValuee=dscc.noDataValue
myArraySumm = SctAgl.sum(1)
myArraySumm.shape = (SctAgl.shape[0],1)
myArrayPercc = (SctAgl * 1.0)/ myArraySumm
newRasterr = arcpy.NumPyArrayToRaster(myArrayPercc,lll,dscc.meanCellWidth,dscc.meanCellHeight,noDataValuee)
arcpy.DefineProjection_management(newRasterr, srr)
OutRaster = os.path.join(outdir,'MOD022222.{0}.tif'.format("SctAgl"))
newRasterr.save(OutRaster)
# # RCR for TOA_BAND_1 and TOA_BAND_2
# TOA_BAND_1
dsc=arcpy.Describe(toa_b1)
sr=dsc.SpatialReference
ext=dsc.Extent
ll=arcpy.Point(ext.XMin,ext.YMin)
noDataValue=dsc.noDataValue
toa_b1_na = arcpy.RasterToNumPyArray(toa_b1)
B1_ROD = (0.00864 + (0.0000065)) * (0.67)**(-(3.916 + (0.074 * 0.67)+ (0.05/0.67)))
Pr1 = (3.0/16.0*np.pi) * (1 + (np.cos(SctAgl) * np.cos(SctAgl)))
wr1 = 1.0
RayRef_B1 = (wr1 * B1_ROD * Pr1 ) / (4.0 * (cosSolZ * cosSenZ))
myArray = toa_b1_na - RayRef_B1
myArraySum = myArray.sum(1)
myArraySum.shape = (myArray.shape[0],1)
myArrayPerc = (myArray * 1.0)/ myArraySum
newRaster = arcpy.NumPyArrayToRaster(myArrayPerc,ll,dsc.meanCellWidth,dsc.meanCellHeight,noDataValue)
arcpy.DefineProjection_management(newRaster, sr)
OutRaster = os.path.join(outdir,'MOD022222.{0}.tif'.format("TOA_B1"))
# toa_b2.save(OutRaster)
newRaster.save(OutRaster)
print arcpy.GetMessages()
print 'finished run: %s\n\n' % (datetime.datetime.now() - start)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi BIJOY GAYEN ,
There is no option, without conversion (Raster To Numpy and then Numpy To Raster) to write the code for raster data analysis directly use cos, sin, subtract, plus, minus, division function, etc. I am totally confused to write this code. Give any solution regarding this issue
Have a look at these functions that are available in Spatial Analyst:
- http://desktop.arcgis.com/en/arcmap/latest/tools/spatial-analyst-toolbox/cos.htm
- http://desktop.arcgis.com/en/arcmap/latest/tools/spatial-analyst-toolbox/sin.htm
- http://desktop.arcgis.com/en/arcmap/latest/tools/spatial-analyst-toolbox/minus.htm
- http://desktop.arcgis.com/en/arcmap/latest/tools/spatial-analyst-toolbox/plus.htm
- http://desktop.arcgis.com/en/arcmap/latest/tools/spatial-analyst-toolbox/divide.htm
- http://desktop.arcgis.com/en/arcmap/latest/tools/spatial-analyst-toolbox/power.htm
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hello Xander Bakker sir, sorry for the late reply. I am confused to write the code using your mentioned function. Can you write one example code with my code?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
My example uses the in_memory workspace only because I'm writing summary tables there and they are relatively small. My initial approach was to write everything to disc in a file geodatabse; for me that was more overhead than I needed. Processing rasters data is a different animal altogether. Using in_memory is great for speed, but if it gets consumed, things have a tendency to go south....
- « Previous
- Next »
- « Previous
- Next »