- Home
- :
- All Communities
- :
- Products
- :
- ArcGIS Image Server
- :
- ArcGIS Image Server Questions
- :
- Rename raster bands in ArcGIS 10.x
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi all,
I have imported a multiband raster in .tif format into a geodatabase using ArcGIS 10.7 and want to rename the bands to something more descriptive. However, I can't find out how to do this easily.
Does anyone know how to rename bands in ArcGIS 10.x?
Solved! Go to Solution.
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi Wade,
to answer this one first: In a MosaicDataset you can create "custom" band definitions: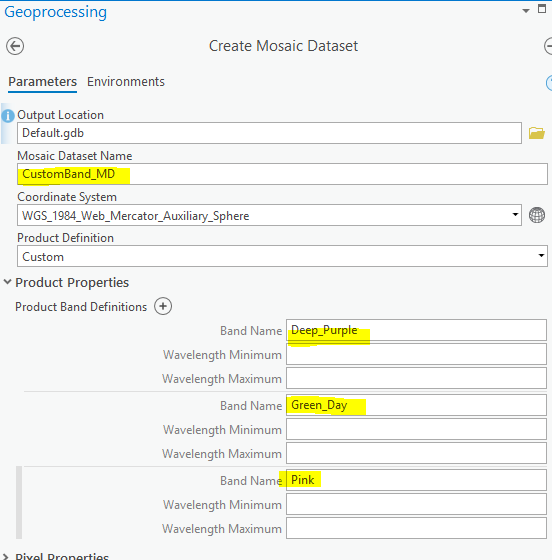
which lead to these custom band names being accessible in UI ...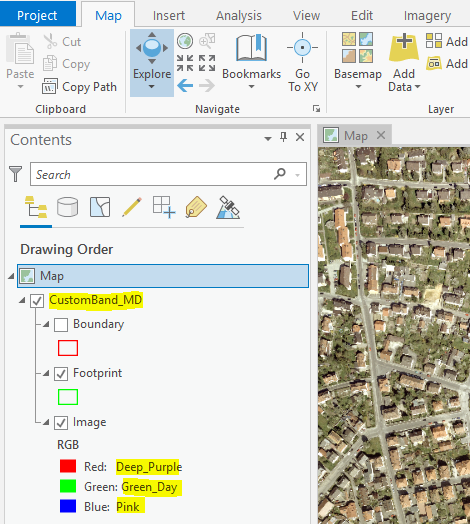
Raster Functions ...
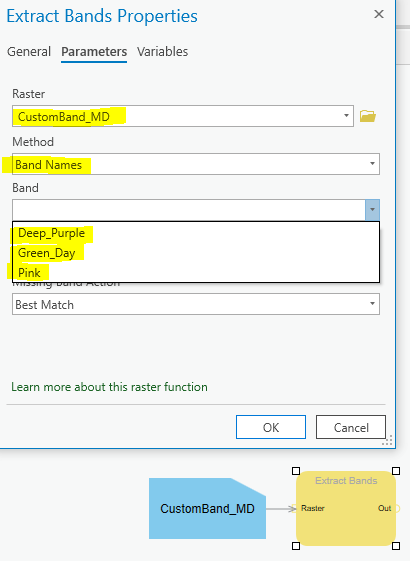
Python code ..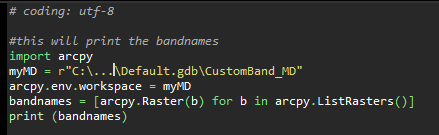
The gp-tool "arcpy.SetMosaicDatasetProperties_management()" will allow you to also SET those definitions, but personally I never used that
G.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Best: Create a MosaicDataset (where in the definition of the Mosaic, you can define Band Names, bandwidth, ...) and then load your dataset into it ... also will allow you to define proper processing templates for suitable band combinations
G.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Thanks for the reply. I can create a new Mosaic Dataset, but still not sure how to rename bands. Could you provide an example of how to create a four band raster with the bands being named "This","Does","Not","Work"? I have tried to do it through multiple combinations of values.
As a side not, this seems like a very difficult way to rename bands. It should be as easy as something like raster.bandnames = ['a','b','c']. Just complaining.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi Wade,
to answer this one first: In a MosaicDataset you can create "custom" band definitions:
which lead to these custom band names being accessible in UI ...
Raster Functions ...

Python code ..

The gp-tool "arcpy.SetMosaicDatasetProperties_management()" will allow you to also SET those definitions, but personally I never used that
G.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
This is not a solution and nothing has been solved. Where is the Arcpy equivalent for the GDAL 'RasterBand.SetDescription(BandName)'
If I have 1300 images you are essentially asking me to rewrite all of the raster data into a mosaic format, in my case 1.3TB of data just to alter 5 to 6 charters per image; this is not practical and not a solution. This is very easy to do in ENVI and ERDAS. Is there a solution that is effective for batch processing?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
One can right-click on each band name in Arc Catalog and rename bands that way. So how can one do this using arcpy? There has to be a way to access the band names using arcpy.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Following the trail that WadeWall pointed to, I wrote a small Python function that can fix this problem.
I wrote this thinking of a rasterio.DatasetReader object being passed but can be easily adapted to gdal
import os
def writePAMfile(img, descriptions=None):
pam_path = os.path.abspath('.'.join([img.files[0], 'aux.xml']))
n_bands = img.count
if isinstance(descriptions, list):
if not len(descriptions) == n_bands:
raise ValueError("You must pass one description string for every band")
elif descriptions is None:
descriptions = img.descriptions
else:
raise TypeError("The argument 'descriptions' must be a list of strings or None")
with open(pam_path, 'w') as pam:
pam.write("<PAMDataset>\n")
for i in range(n_bands):
if descriptions[i] is None:
pam.write('\t<PAMRasterBand band="{0}">\n\t\t<Metadata>\n\t\t\t<MDI key="BandName">None</MDI>\n\t\t</Metadata>\n\t</PAMRasterBand>\n'.format(str(i+1)))
else:
pam.write('\t<PAMRasterBand band="{0}">\n\t\t<Description>{1}</Description>\n\t\t<Metadata>\n\t\t\t<MDI key="BandName">None</MDI>\n\t\t</Metadata>\n\t</PAMRasterBand>\n'.format(str(i+1), descriptions[i]))
pam.write("</PAMDataset>\n")
In this function you can pass the list of band_names to use, but if those are already set using the appropriate gdal or rasterio method you can leave it blank and will read this information from your raster
Once that file is written, loading the raster in ArcMap or ArcCatalog will show the appropriate names
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Or if you would like a bit more refined functionality, like preserving already existing tags, removing descriptions and so on
import os
import xml.etree.ElementTree as ET
def find_xml_element(parent_element, tag, attribs=None):
options = parent_element.findall(tag)
if len(options) == 0:
return None
else:
if attribs is None:
return options[0]
for o in options:
if o.attrib == attribs:
return o
else:
return None
def writePAMfile(img, descriptions=None):
n_bands = img.count
if isinstance(descriptions, list):
if not len(descriptions) == n_bands:
raise ValueError("You must pass one description string for every band")
elif descriptions is None:
descriptions = img.descriptions
else:
raise TypeError("The argument 'descriptions' must be a list of strings or None")
pam_path = os.path.abspath('.'.join([img.files[0], 'aux.xml']))
if os.path.isfile(pam_path):
tree = ET.parse(pam_path)
root = tree.getroot()
if root.tag != 'PAMDataset':
raise RuntimeError("a XML file {0} exists but is not recognized as a valid PAM dataset".format(pam_path))
else:
for n, d in enumerate(descriptions):
band = find_xml_element(root, "PAMRasterBand", {'band':str(n + 1)})
if band is None:
band = ET.SubElement(root, "PAMRasterBand", band=str(n+1))
meta = band.find("Metadata")
if meta is None:
meta = ET.SubElement(band, 'Metadata')
mdi = find_xml_element(meta, "MDI", {'key':'BandName'})
if mdi is None:
ET.SubElement(meta, "MDI", key="BandName").text = str(d)
else:
mdi.text = str(d)
desc = band.find("Description")
if d is not None:
if desc is None:
ET.SubElement(band, "Description").text = d
else:
desc.text = d
else:
if desc is not None:
band.remove(desc)
else:
root = ET.Element("PAMDataset")
for n, d in enumerate(descriptions):
band = ET.SubElement(root, "PAMRasterBand", band=str(n+1))
meta = ET.SubElement(band, 'Metadata')
if isinstance(d, str):
ET.SubElement(meta, "MDI", key="BandName").text = d
ET.SubElement(band, "Description").text = d
elif d is None:
ET.SubElement(meta, "MDI", key="BandName").text = "None"
else:
raise ValueError("Descriptions must be a str or None")
tree = ET.ElementTree(root)
tree.write(pam_path)