- Home
- :
- All Communities
- :
- Products
- :
- ArcGIS Pro
- :
- ArcGIS Pro Questions
- :
- Re: unexpected truncation in my text field ("Table...
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
unexpected truncation in my text field ("Table to Table" GP)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
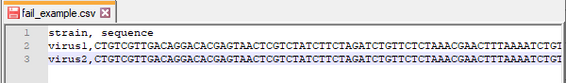
I am using the "Table to table" GP to input the following csv to gdb for genetic data.
the sequence of virus1 has length about 31000
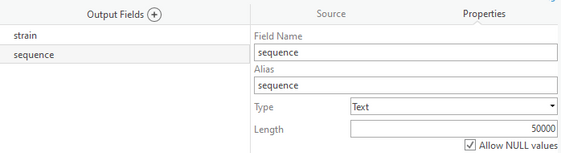
The default text length in the tool is 8000 and I amended it to 50000
The import ran successfully and I added a field for finding the maximum sequence length.
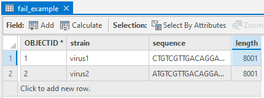
However, when I looked in the attribute table.
The import string is truncated to length of 8001 which originally should be ~31000.
Question: How do I fix it?
I have attached the required testing file.
Thank for anyone that can help with this issue.
Solved! Go to Solution.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
@Felix10546 if you are good with numpy, then you can just use
# -- set dtype to string rather than unicode
dt = np.dtype([('strain', 'S6'), ('sequence', 'S50000')])
ar2 = np.genfromtxt(f, dtype=dt, delimiter=",", names=True, autostrip=True, encoding='utf-8')
ar2.dtype
dtype([('strain', 'S6'), ('sequence', 'S50000')])
fc1 = r"C:\arcpro_npg\npg\Project_npg\tests.gdb\strain2"
NumPyArrayToTable(ar2, fc1)
If you use unicode strings it doubles your field widths, if you stick with string S, the field widths are retained
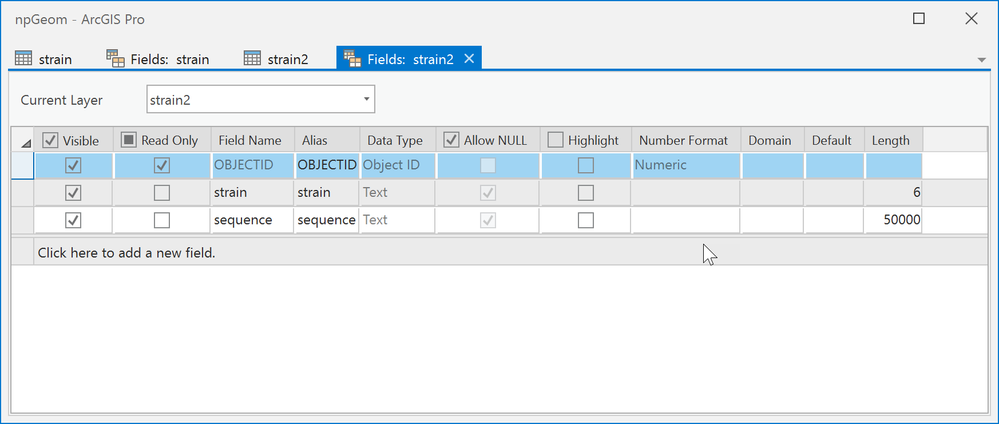
Here is the fields view of the data you posted.
BTW NumPyArrayToTable skips all the hoops to get a table if you are working with csv data
... sort of retired...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
I don't know I can directly interact with data in gdb with numpy. The data is not in my hand now. Let me try tmr.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Yah, the script work and the length of the table can be controlled in the generating process by this way. Thank DanPatterson.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
glad it worked out
... sort of retired...
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Light reading for the holidays
ExtendTable—ArcGIS Pro | Documentation
FeatureClassToNumPyArray—ArcGIS Pro | Documentation
NumPyArrayToFeatureClass—ArcGIS Pro | Documentation
NumPyArrayToTable—ArcGIS Pro | Documentation
TableToNumPyArray—ArcGIS Pro | Documentation
and I have lots of blog post in the python blog
... sort of retired...
- « Previous
-
- 1
- 2
- Next »
- « Previous
-
- 1
- 2
- Next »