- Home
- :
- All Communities
- :
- Developers
- :
- Python
- :
- Python Questions
- :
- How to extract metadata of feature classes and sav...
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
How to extract metadata of feature classes and save it as a csv, using ISO19139 translator in ArcGIS environment ?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
We are trying to extract the feature_classes metadata and save it as csv. With the help of this code we are extracting Abstract, Title, Purpose, Spatial Ref, Extent without any hiccups. But in addition to that we need "Lineage" info and i don't see any lineage info in xml files converted using "ARCGIS2FGDC.xml" translator. But lineage details are getting exported when ARCGIS2ISO19139.xml translator is used.
The problem is, i couldn't able to extract any of the details (Abstract, Lineage, Title etc..) from the output xml translated using ARCGIS2ISO19139.xml. This is the code i am using
import os
import arcpy
import csv
from xml.etree.ElementTree import ElementTree
from arcpy import env
arcpy.env.overwriteOutput = True
def inventory_data(workspace, datatypes):
for path, path_names, data_names in arcpy.da.Walk(
workspace, datatype=datatypes):
for data_name in data_names:
yield os.path.join(path, data_name)
AGSHOME = arcpy.GetInstallInfo("Desktop")["InstallDir"]
translatorpath = AGSHOME + "Metadata\\Translator\\ARCGIS2ISO19139.xml"
#These two variables need to be updated each time this script is run on a new workspace
outfile = r"D:\QAQC\Metadata\GIS_Data_Inventory.csv"
xmlfile = r"D:\QAQC\Metadata\GIS_Data_Inventory.xml"
with open (outfile, 'wb') as csvfile:
csvwriter = csv.writer(csvfile)
#You need to update workspace in the line below
for feature_class in inventory_data(r"D:\QAQC\Metadata\Test", "FeatureClass"):
try:
desc = arcpy.Describe(feature_class)
sr = desc.spatialReference
arcpy.ExportMetadata_conversion(feature_class, translatorpath, xmlfile)
tree = ElementTree()
tree.parse(xmlfile)
try:
title = tree.find ("identificationInfo/MD_DataIdentification/citation/CI_Citation/title").text
except:
title = "No Title"
try:
abstract = tree.find ("identificationInfo/MD_DataIdentification/abstract").text
except:
abstract = "No Abstract"
try:
lineage = tree.find ("dataQualityInfo/DQ_DataQuality/lineage/LI_Lineage/statement").text
except:
lineage = "No Lineage"
csvwriter.writerow([desc.path.encode('utf-8'), desc.file.encode('utf-8'), desc.dataType.encode('utf-8'), sr.name.encode('utf-8'), purpose.encode('utf-8'), title.encode('utf-8'), abstract.encode('utf-8')])
except Exception:
e = sys.exc_info()[1]
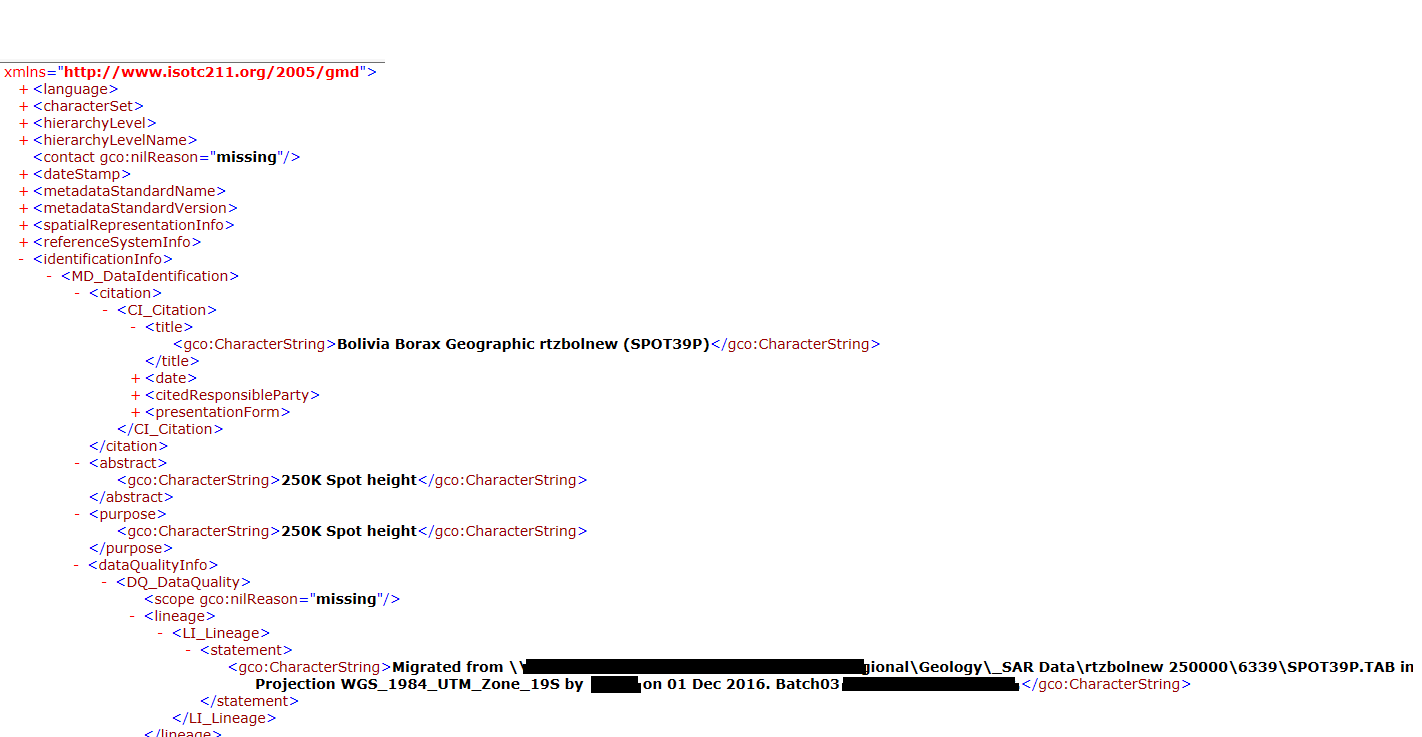
print(e.args[0])All i am getting in the output csv is "No Title, No Abstract, No Lineage". But the output xml file has these parameters (image attached below).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Could you explain what you are trying to do here? Metadata is very complex structured data (for example, the lineage is a collection of objects (one or more), each that can have different properties). CSV by contrast is a simple text table.
Could you show what your goal is here, what your output text file should look like?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
We have to QAQC 100's of feature_classes and one of the checks is to check the metadata description. So we are trying to extract Abstract, Title, Extent, Lineage Statement of the feature_classes and save it as a csv file, so that we can easily check the things using filter options in the csv(excel) sheet.
Using the ARCGIS2FGDC.xml tranlstor, we can't able to extract "Lineage -> Statement". But ISO19139.xml is doing the job. The problem is, the code i've mentioned in the question doesn;t extract any of the details. All we are getting is a empty csv file.
Output text should be like (w.r.t the image attached):
250K Spot height, Bolivia Borax Geographic rtzbolnew (SPOT39P), Extent details, Migrated from ________________ (Lineage Statement)
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
perhaps you could format your code so that potential errors could examined... it would separate those from other problems /blogs/dan_patterson/2016/08/14/script-formatting
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Done. Thanks for the link
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Do you get the same result when you run the Geoprocessing Toolbox? (note: I had the same nothing/no output result as you when I simply ran the existing GP tool from ArcCatalog).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
Hi
Once you have sensible output from Export Metadata you could try the Data Interoperability extension to create CSV.
A free trial is available. You will probably need to inspect the structure of the XML to get the paths right. If you need help ask your account manager to assist.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
I don't use "import csv" , but I'm wondering if you need to close you csv file. Here is a sample of how I write to a csv file, including added a header record first. This does an inventory on feature classes, but is similar to your code. As Dan mentioned, make sure to format you code in the future. It is hard for others to debug since python spacing is critical for looping, etc. My close statement is on line 51 below
# snippet of writing to csv file...this inventories feature classes
import arcpy
import os
theWorkspace = r"c:\__temp"
outFile = "FCInventory"
outFileCSV = os.path.join(theWorkspace, ("{0}.csv".format(outFile)))
csvFile = open(outFileCSV, 'w')
# my headers
csvFile.write("FType, FCname, recCount, FullPath\n")
def inventory_data(workspace, datatypes):
for path, path_names, data_names in arcpy.da.Walk(
workspace, datatype=datatypes):
if "tic" in data_names:
data_names.remove('tic')
for data_name in data_names:
fcName = os.path.join(path, data_name)
#print("Show for debug: " + fcName)
if not arcpy.Exists(fcName):
# workaround for raster folder name duplicating
fcName = os.path.dirname(fcName)
desc = arcpy.Describe(fcName)
count = arcpy.GetCount_management(fcName).getOutput(0)
#print("debug, desc it to me: " + desc.dataType)
yield [path, data_name, desc.dataType, count] #, desc]
i = 0
for feature_class in inventory_data(theWorkspace, "FeatureClass"):
if i == 0:
print("{0}".format(feature_class[0])) #print(' ' + feature_class[0])
outText = (" {0}\n".format(feature_class[0])) # ' ' + feature_class[0] + '\n'
path0 = feature_class[0]
i =+ 1
elif not path0 == feature_class[0]:
print(' ' + feature_class[0])
outText = ' ' + feature_class[0] + '\n'
i = 0
print(" " + feature_class[2] + ": " + feature_class[1])
outText = (" " + feature_class[2] + ": " + feature_class[1] + '\n')
csvFile.write("{0},{1},{2},{3}\n".format(feature_class[2], feature_class[1], feature_class[3], feature_class[0]))
#csvFile.write("{},{}, {}\n".format(feature_class[2], feature_class[1], feature_class[0]))
csvFile.close()
print( "File {0} is closed? {1}".format(outFileCSV, str(csvFile.closed)))
print('!!! Success !!! ')
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
It may be easier to find out what's wrong if you don't just silently pass errors, but report what the error was (for debugging purposes):
try:
title = tree.find ("identificationInfo/MD_DataIdentification/citation/CI_Citation/title").text
except Exception as msg:
print("Error parsing for {}:".format(feature_class))
print(str(msg))
title = "No Title"