- Home
- :
- All Communities
- :
- Developers
- :
- Python
- :
- Python Blog
- :
- Euclidean distance, allocation and other stuff...
Euclidean distance, allocation and other stuff...
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
Your standard fare in raster world
Fill in some holes … 'buffer' a line... allocate space to stuff in space... identify unique regions
Fancy names. Nibble, Euclidean distance, Euclidean allocation, Regiongroup
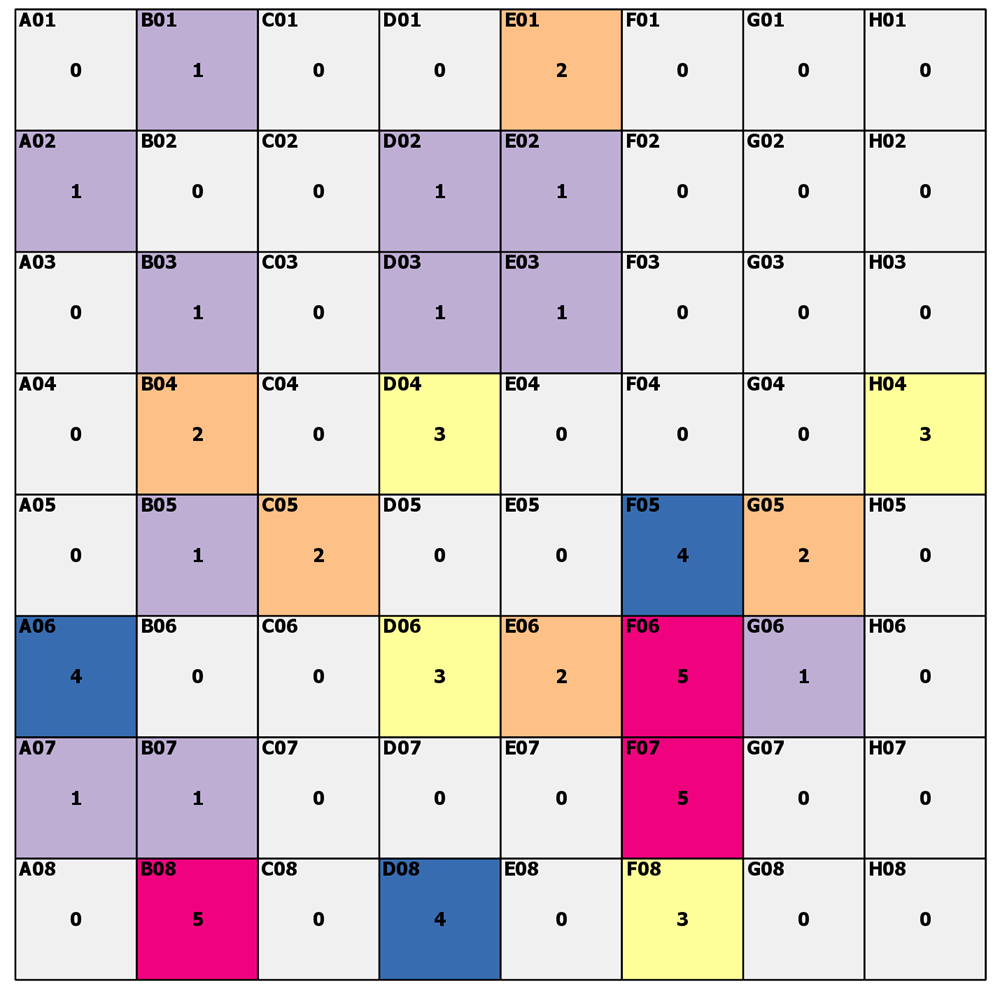
---- (1) The task ----
Start with a raster or array. Fancy labels in the top left, some random-ish color scheme with values noted in the middle.
Now, zero ( 0 ) we will say is nodata. The other numbers represent some class value.
Fill in the 'gaps'... aka, nodata, with the value of a cell with data based on the closest distance. Euclidean, crow flies, for our purposes, but it need not be.
Go!
What did you get for cells A04, H01 and H02? What about cell D07 and H08?
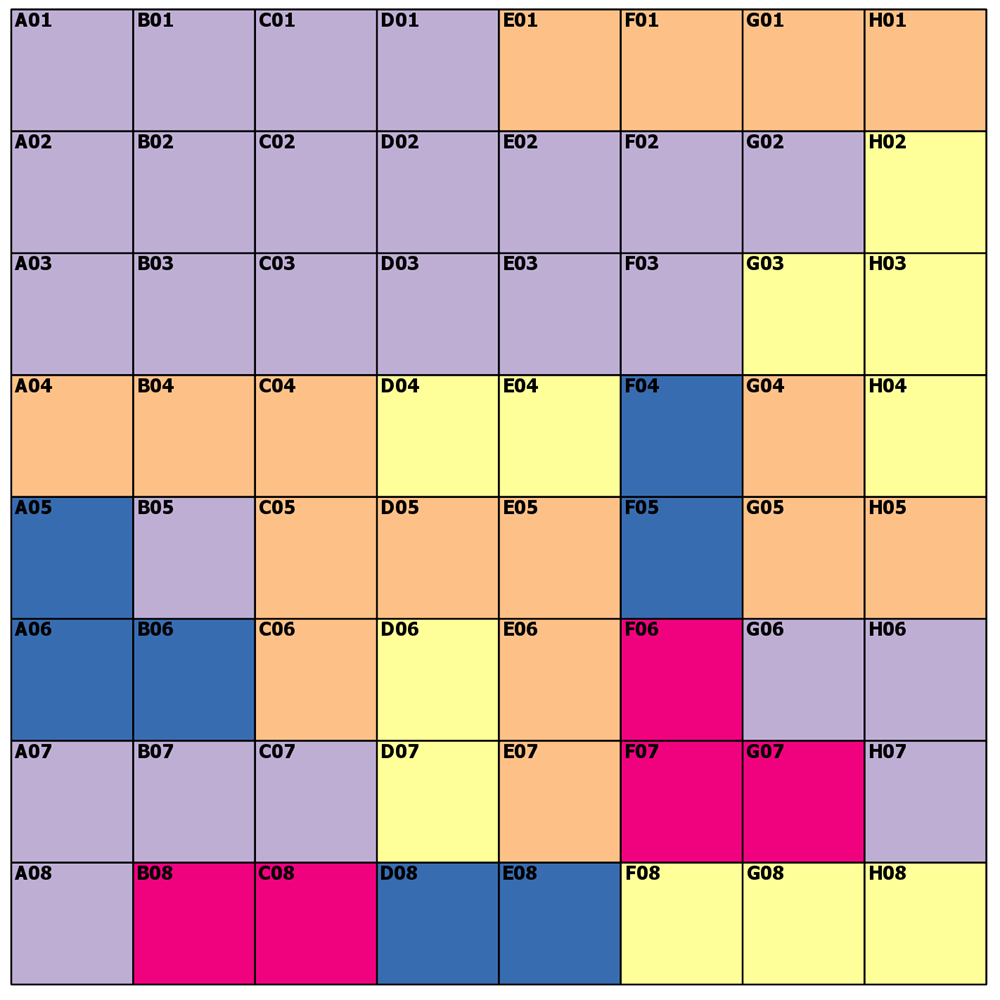
---- (1) The reveal ----
Let's see how we did
A04 - 2 can't be 1 because the diagonal to '1' is 1.414*cell width, so 2 it is
H01 - 2 could have been a 2 or a 3, because both are 3 cells away from cells with values
H02 - 3 no brainer
D07 - 3 could have been 3 or 4, but 3 wins
H08 -3 3 is 2 cells away and 1 and 5 are a greater distance on an angle
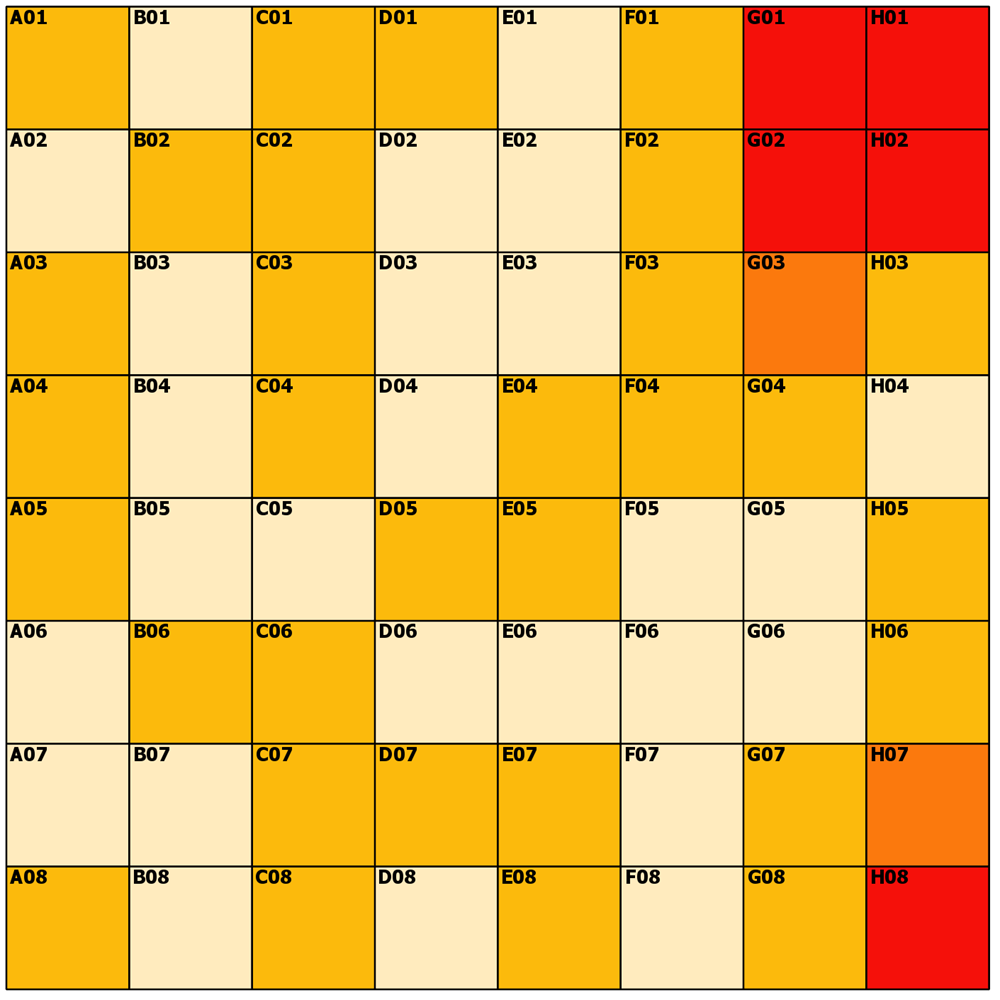
---- (3) The distances ----
Pretty boring and obvious... but for completeness.
I don't do maps, but the dominant colors are 0 or root 2 as you can see from the spiffy ArcGIS Pro symbology tab.So no big surprises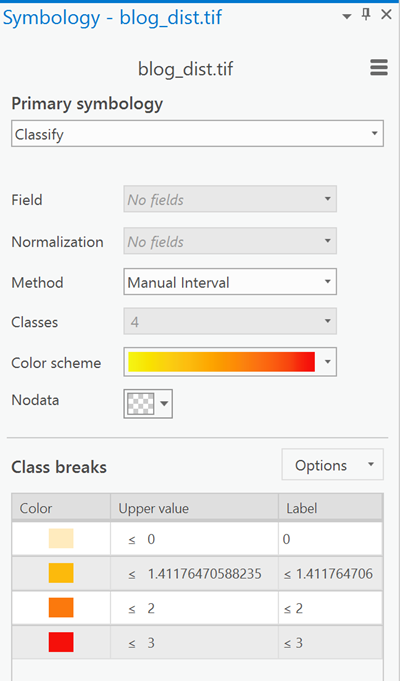
---- (4) Allocation around linear features ----
Yes, that is possible too, sort of like a variable buffer, trade area, but currently just a simple Euclidean spatial allocation.

---- (5) The references ----
NumPy/SciPy/ plus Arcpy stuff solution is what I used
import sys
import numpy as np
from scipy import ndimage as nd
from arcpy.geoprocessing import env
from arcpy.arcobjects import Point
from arcgisscripting import NumPyArrayToRaster, RasterToNumPyArray
env.overwriteOutput = True
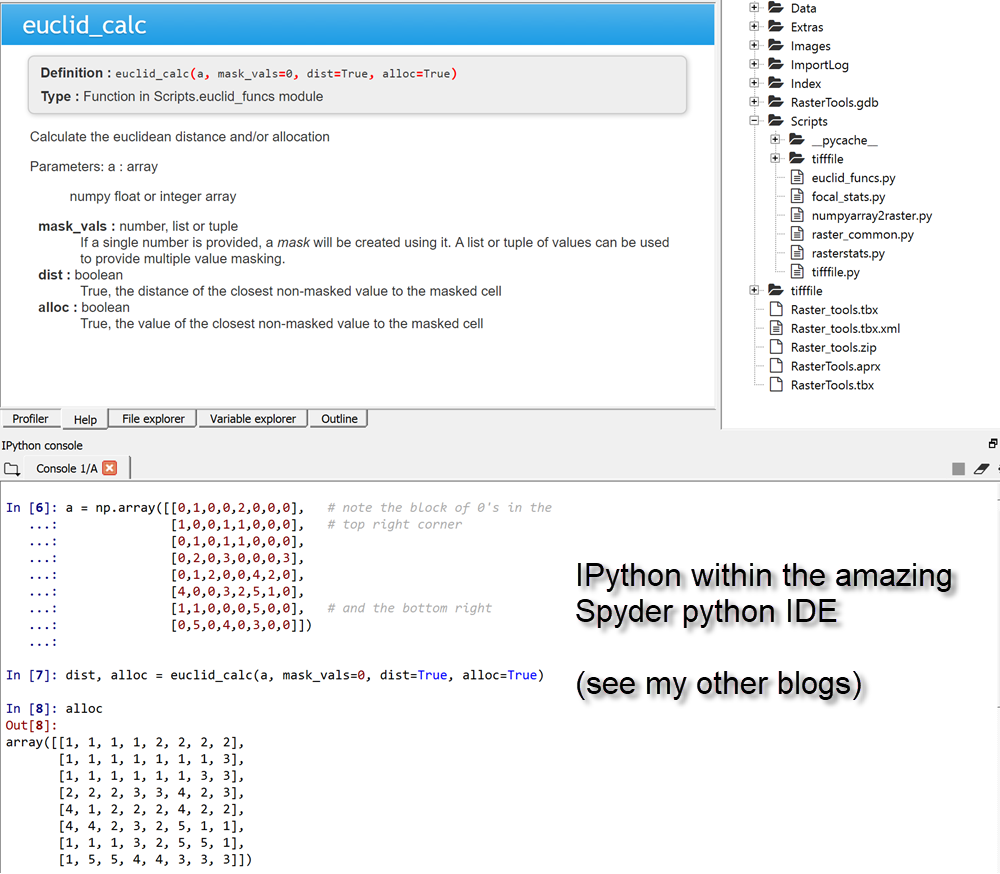
def euclid_calc(a, mask_vals=0, dist=True, alloc=True):
"""Calculate the euclidean distance and/or allocation
Parameters:
a : array
numpy float or integer array
mask_vals : number, list or tuple
If a single number is provided, a `mask` will be created using it. A
list or tuple of values can be used to provide multiple value masking.
dist : boolean
True, the distance of the closest non-masked value to the masked cell
alloc : boolean
True, the value of the closest non-masked value to the masked cell
"""
if not dist:
dist = None
if not alloc:
alloc = None
m = np.isin(a, mask_vals).astype('int')
dist, idx = nd.distance_transform_edt(m, return_distances=True,
return_indices=True)
alloc = a[tuple(idx)]
return dist, alloc
# ----------------------------------------------------------------------
# .... final code section
if len(sys.argv) == 1:
testing = True
r_in = r"C:\GIS\A_Tools_scripts\Raster_tools\Data\poly.tif"
a = RasterToNumPyArray(r_in)
dist, alloc = euclid_calc(a, mask_vals=0, dist=True, alloc=True)
else:
testing = False
r_in = sys.argv[1]
r_out = sys.argv[2]
LLx = sys.argv[3]
LLy = sys.argv[4]
cell_sze = sys.argv[5]
a = RasterToNumPyArray(r_in)
dist, alloc = euclid_calc(a, mask_vals=0, dist=True, alloc=True)
#
# specify a, dist or alloc below... this could be a tool choice if needed
#
r0 = NumPyArrayToRaster(a, Point(LLx, LLy), cell_sze)
r0.save(r_out)Of course, when run from within the amazing Spyder python IDE, you can simply skip saving to output raster if needed, or try various other options from with the scipy slate other than the distance_transform_edt
On to other "special analyst" tools soon.
You must be a registered user to add a comment. If you've already registered, sign in. Otherwise, register and sign in.