- Home
- :
- All Communities
- :
- Products
- :
- Imagery and Remote Sensing
- :
- Imagery Questions
- :
- Re: UnetClassifier with ignore_classes
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
UnetClassifier with ignore_classes
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I have noticed some interesting results when using UnetClassifier with ignore_classes.
First if you only have 3 classes, such as with this code
data = prepare_data(data_path, batch_size=32, chip_size=224, num_workers=0)
data.classes
The output is : ['NoData', '1', '2']
model = UnetClassifier(data,backbone='resnet34', ignore_classes=[0])
The above line will error with the complaint:
Exception: `ignore_classes` parameter can only be used when the dataset has more than 2 classes.
But the stats.txt file shows 3 classes. Snippet below
images = 134 *12*224*224
features = 2279
features per image = [min = 1, mean = 17.01, max = 67]
classes = 3
cls name cls value images
0 0 125
1 1 120
2 2 117
Seems like a bug to me, but the workaround is to create a fake class (say class 10) and re-export the tiles again and replace the above code with:
model = UnetClassifier(data,backbone='resnet34', ignore_classes=[0,10])
I have also noticed that when using ignore_classes parameters the ‘dice’ statistics in the training data output is usually pretty poor, such as 50-60.
epoch train_loss valid_loss accuracy dice time
…
568 0.541224 0.451169 0.823589 0.658838 00:04
569 0.539253 0.448183 0.822532 0.660908 00:04
As opposed to when there is no 0 class (i.e. [Nodata]) you will normally get high 90 dice results. I assume this is most likely because the dice algorithm is calculating on the [NoData] areas, when I don’t think it should. So therefore results will be highly dependent on the amount of [NoData] areas you have.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
More on this, even when training using this command
model = UnetClassifier(data,backbone='resnet34', ignore_classes=[0,10])
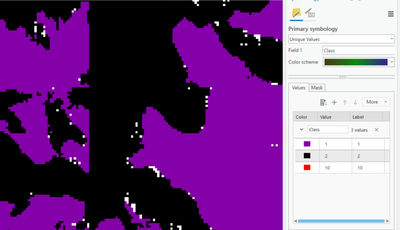
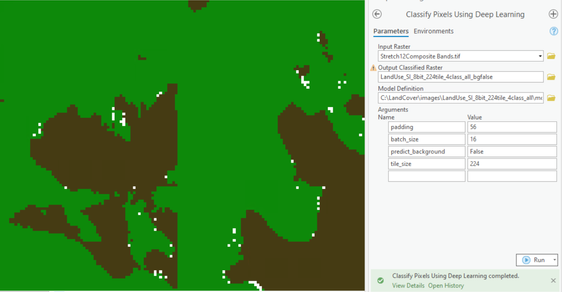
and then using the ClassifyPixelsUsingDeepLearning, the 0 (NoData) is still appearing, see the white dots below
When I click on the white dots the Class is 0.
Anyone have any ideas on this?
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
This is expected, segmentation models work with Nodata (Fixed) + Classes(In training data). I am afraid there is no way we can get rid of that in training.
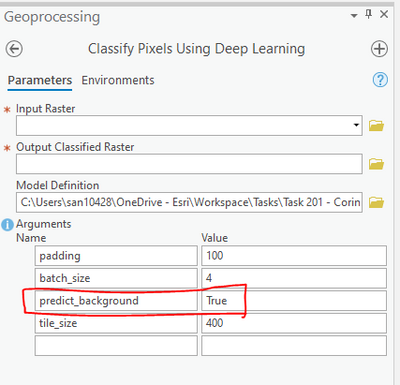
However while deploying the trained model there is an option in the tool to predict background, you can try setting it to False.
Thanks,
Sandeep
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Thanks Sandeep for your help, I haven't had much luck even though predict_background is False.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
@TimG I believe convention is for the value of classvalue to start at 1, and not 0. I believe I've seen documentation to that effect, but I cannot find it at this moment. As a result of that, what you might be seeing is that your 0 class (if that is a legitimate value) is being combined or overwritten by NoData, because 0 is the conventional value for NoData. A NoData class should not show up in stats.txt, in my experience.
When I ignore NoData, as you did, I do get the occasional pixel that comes up NoData (as you did in your reply), though there are significantly less of them than if I don't ignore NoData. I think Sandeep's suggestion of predict_background works better when the NoData class is still included, whereas I haven't seen much difference in the result of predict_background True vs False when NoData was ignored.